A group of mutants suitable for classification and determination of patient prognosis in liver cancer histology
A technology for prognosis and mutation of liver cancer, applied in biochemical equipment and methods, analytical materials, scientific instruments, etc., can solve problems such as unclear effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0181] Example 1 Detection of LLGL1 mutation in liver cancer
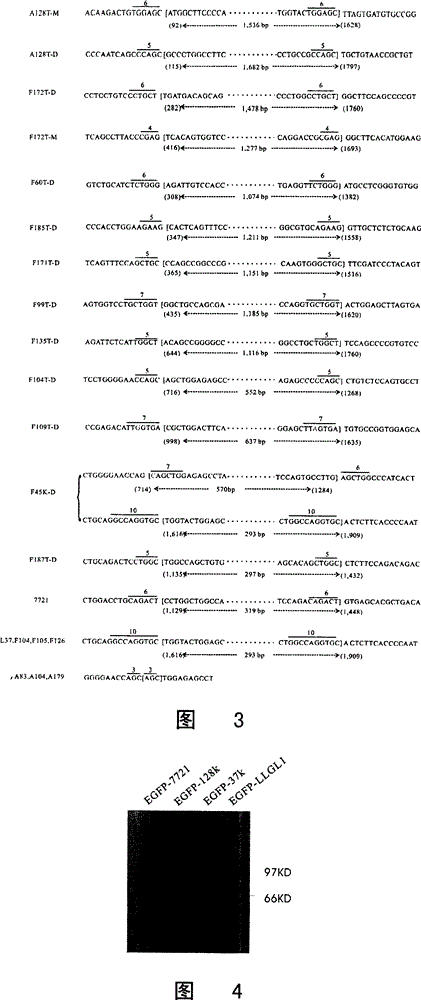
[0182] Using the total RNA extraction reagent Trizol (Invitrogen Company) according to the operating instructions, extract 78 pairs of patients' liver cancer and paracancerous tissues and 5 strains of liver cancer cell lines (hepG2, huh7, SKhep1, SMMC-7721, BEL-7404, from the Chinese Academy of Sciences According to the total RNA, the total RNA was reacted with RT at 25°C for 5 minutes, 42°C for 60 minutes, and 70°C for 15 minutes. Four primers (P1: 5'-GGAATTCCGCAAGATGATGAAGTTTCG-3' (SEQ ID NO: 13) and P2: 5'-GACTGGCGCAGTGACTTCTTG-3' (SEQ ID NO: 14); P35'-ACTCTTCACCCCAATGACT-3' (SEQ ID NO: 15) and P45'-GCTCTAGAGATGCAACAGGCGTGGGCCT-3' (SEQ ID NO :16)) (eg figure 1 Shown in A) LLGL1 was amplified by two-stage PCR, and the cDNA of these samples was obtained from the reaction, the amplified PCR products were detected by 1% agarose gel, and the recovered PCR products were sequenced and analyzed (Boya Company). The in...
Embodiment 2
[0193] Example 2 Preparation of Three LLGL1 Mutants
[0194] Using primers 5'-CGGAATTCCAAGATGATGAAGTTTCGGTTC-3' (SEQ ID NO: 7) and 5'-GCTCTAGAT GATGGCCACGCTGACCGCC-3' (SEQ ID NO: 8), the cDNA of 7721 cells was amplified by PCR, and the amplified product was double-treated by EcorI enzyme and XbaI enzyme. After digestion, clone on the plasmid pEGFP-C1 (purchased from Invitrogene Company) to obtain the recombinant plasmid pEGFP-C1-7721 ( Figure 4 LLGL1-7721-EGFP in ). Two pairs of primers P1: 5'-GGAATTCCGCAA GATGATGAAGTTTCG-3, (SEQ ID NO: 9) and P2: 5'-GACTGGCGCAGTGACTTCTTG-3' (SEQ ID NO: 10); P3 5'-ACTCTTCACC CCAATGACT-3' (SEQ ID NO: 10) were used respectively. ID NO: 11) and P4 5'-GCTCTAGAGATGCAACAGGCG TGGGCCT-3' (SEQ ID NO: 12), PCR amplified 37k and 128k abnormal transcripts in two patient specimens respectively, and the amplified products were double-passed by EcorI enzyme and SmaI enzyme Digest the 5' (P1 / 2 amplification product) end and the 3' (P3 / P4 amplification prod...
Embodiment 3
[0196] Embodiment 3 detection kit
[0197] A liver cancer prognosis detection kit is prepared, wherein the following primer pair that can amplify the nucleotides from the 92nd to 1627th bases of SEQ ID NO: 1 is deleted: 5'-ACTCTTCACCCCAATGACT-3' (SEQ ID NO: 11); 5'-GCTCTAGAGATGCAACAGGCG TGGGCCT-3' (SEQ ID NO: 12).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com