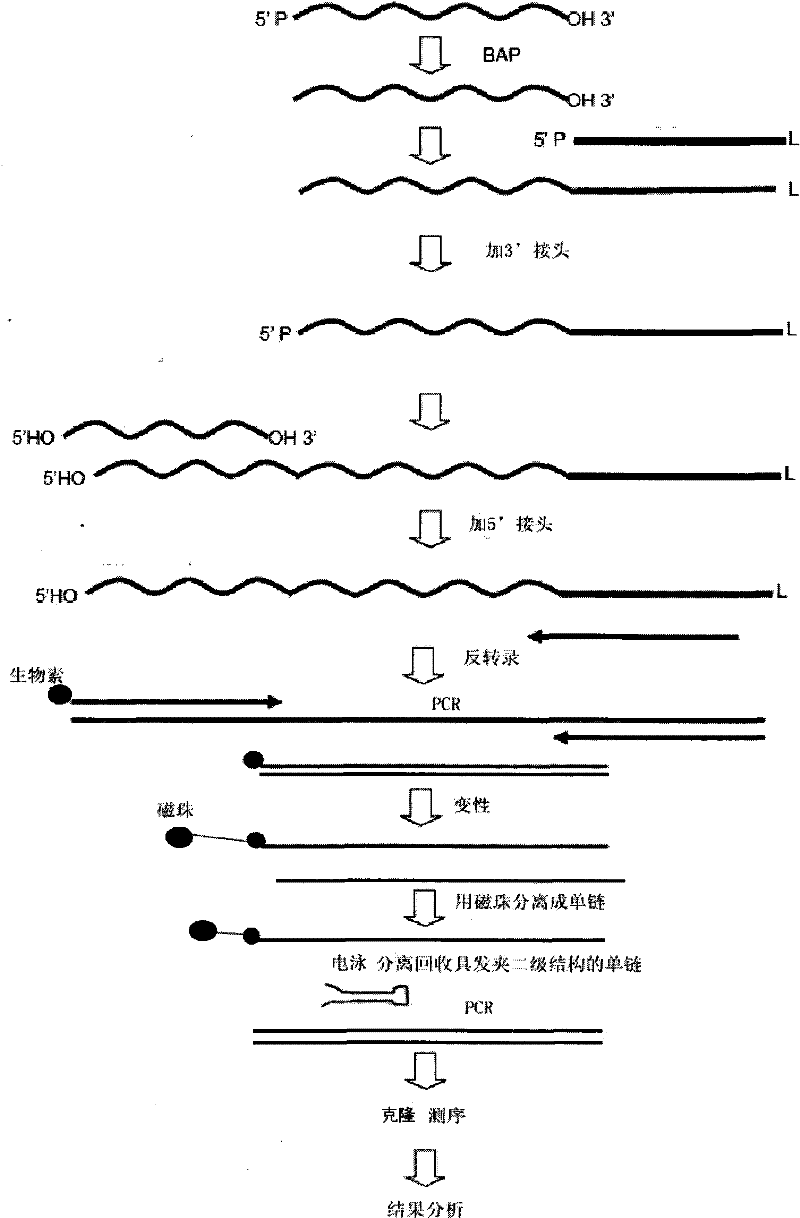
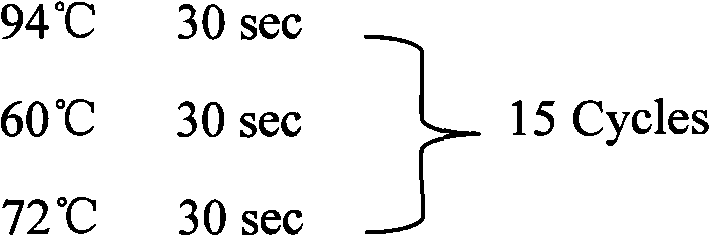
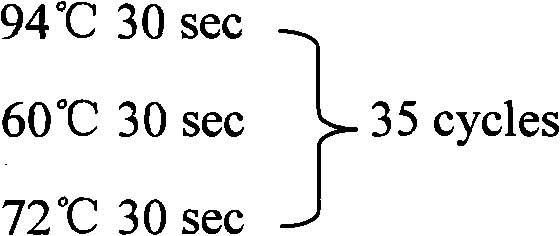
Method for extracting microRNA precursor cDNA from cDNA library synthesized from small RNA
A technology for isolating and precursors, applied in the field of molecular biology, can solve problems such as reduced cloning effect, difficulty in finding miRNA, and many repeated sequences, so as to achieve good separation effect and improve the efficiency of cloning and sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] 1. Small RNA extraction
[0018] A Roche small RNA (<100bp) extraction kit (High Pure miRNA Isolation Kit, Cat. No. 05 080 576 001) was used. The specific operation steps are as follows:
[0019] 1) Add 400 μL 20% binding buffer (Binding Buffer) to a 2 mL empty test tube;
[0020] 2) Add 1-50 mg of muscle tissue sample to the test tube, and break the tissue with a homogenizer;
[0021] 3) Centrifuge the test tube at 14,000 rpm for 2 minutes at 4°C, and then take about 150-188 μL of the supernatant into another new 1.5 mL test tube;
[0022] 4) Add 312μL of Binding Buffer and shake for 3×5s;
[0023] 5) Assemble the filter tube and collection tube in the kit, and suck the mixed solution into the filter tube above;
[0024] 6) Centrifuge at 13000×g for 30s, and collect the liquid in the collection tube below;
[0025] 7) Add 200 μL of Binding Enhancer to the collection tube and shake for 3×5 s;
[0026] 8) Assemble the filter tube and collection tube in the kit, and...
Embodiment 2
[0246] Using this method to study porcine miRNA, the cDNA single strands in two different regions in the gel were separated and recovered, and then cloned and sequenced after recovery to double strands. After the sequencing results were analyzed with BioEdit and RNAfold software, it was found that the ratios of hairpin sequences in the sequencing results of clones in A and B regions were significantly different. Among the 63 sequencing results in area A, almost all of the sequences had a "clover" type secondary structure, and none of them basically met the hairpin type secondary structure standard of miRNA precursors. This shows that most of the small RNAs in this region are not miRNA precursors. In the 45 sequencing results in area B, a total of 5 sequences were found that basically conformed to the secondary structure of the miRNA precursor hairpin, but most of the other sequences still did not meet the definition requirements of the miRNA precursor hairpin secondary structu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com