Nucleic acid aptamer capable of being specifically bound to tubercle bacillus antigen and application thereof
A nucleic acid aptamer and specific binding technology, applied in sugar derivatives, instruments, analytical materials, etc., can solve the problems of difficulty and randomness in obtaining high-affinity aptamers, and achieve low cost and simple preparation , to overcome the effect of low sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
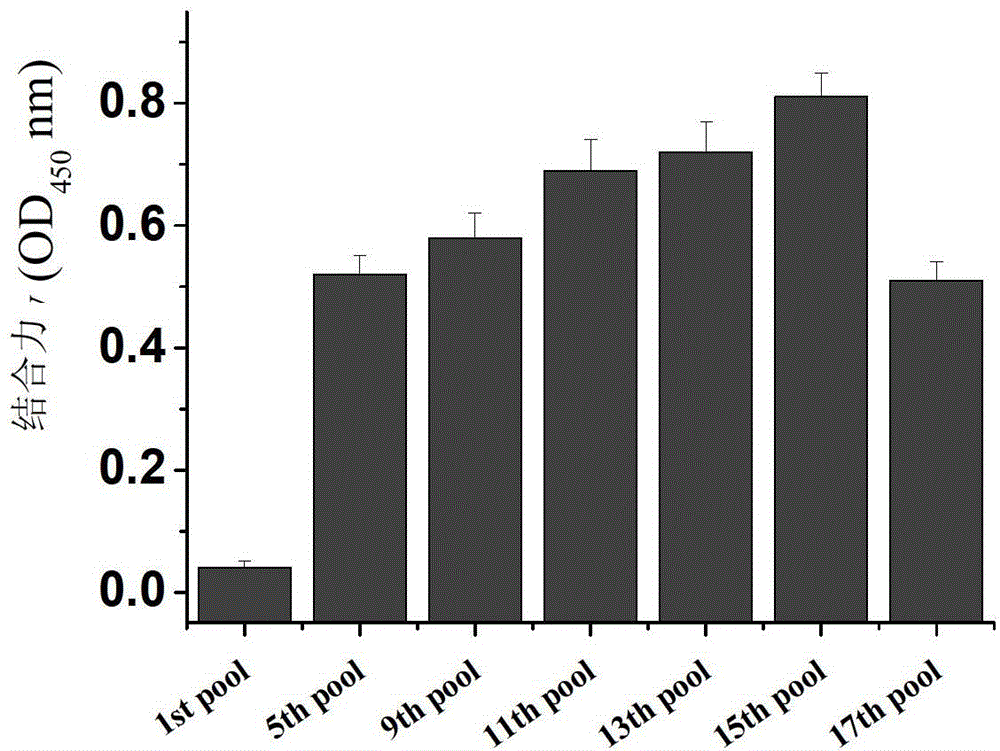
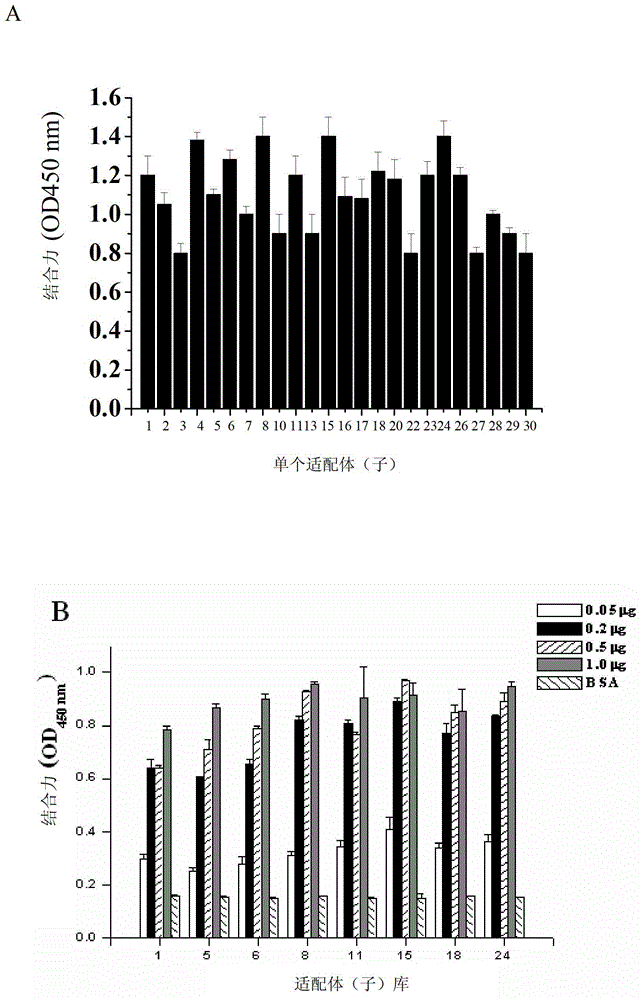
[0031] Example 1 Screening of aptamers
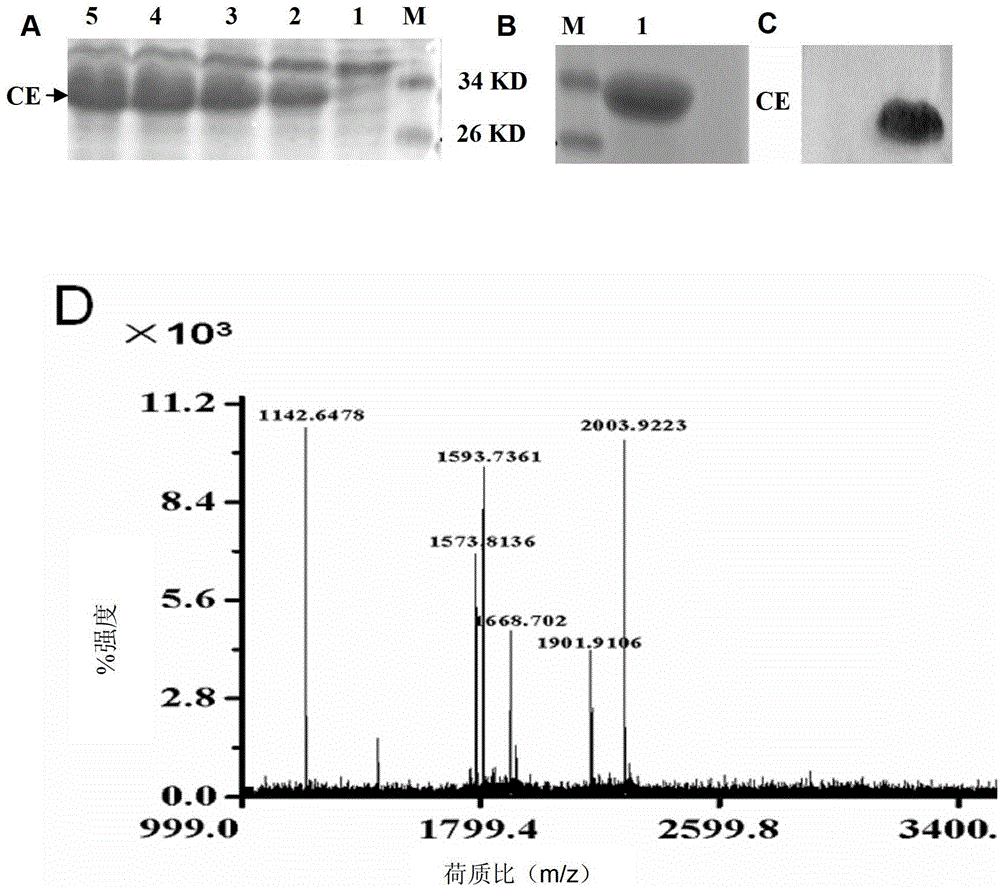
[0032] 1. Purification and identification of CFP10-ESAT6 fusion protein
[0033] Construction of prokaryotic expression plasmid pET28a-CFP10-ESAT6 (or pET28a-CE, Li Juan, Wu Xueqiong, Zhang Junxian, etc. High expression of Mycobacterium tuberculosis CFP10-ESAT6 fusion protein in Escherichia coli[J]. Chinese Journal of Antituberculosis, 2004,26(4):204-208.), adding a linker sequence between CFP-10 and ESAT-6 fragments, the amino acid sequence corresponding to this sequence is GGGGSGGGGSGGGGS, the existence of this sequence can improve the CFP10-ESAT6 fusion protein ( The flexibility between the CFP-10 and ESAT-6 sequences in the CE fusion protein or CE protein for short, thereby fully maintaining their respective natural conformations.
[0034] Transform the correct plasmid into Escherichia coli BL21, from kanamycin resistance (Kan r ) plate to pick a single clone, inoculated in 5ml sterile LB medium with kanamycin added at 1:1000 (try...
Embodiment 2
[0057] Example 2 The sensitivity of antibody-coated aptamer detection sandwich method to detect CE protein
[0058] After the 96-well ELISA plate (Costar), the rabbit anti-CE protein antiserum (or rabbit anti-tuberculosis H37Rv polyantiserum) was diluted at 1:200~1:500 to coat the ELISA plate, and the CE protein and BSA were used Carbonate buffer at pH 9.6 (Na 2 CO 3 1.5g,NaHCO 3 2.9g,Na 2 N 3 0.2g, add water 1000ml, adjust the pH to 9.6) to prepare 10μg / ml, 1μg / ml, 100ng / ml, 10ng / ml, 1ng / ml, 0.1ng / ml, 0.01ng / ml concentration gradient, add 50μl to each well, After incubation overnight at 4°C, wash with PBST 3 times, 2 minutes each time; then add 100 μl blocking solution (0.2% gelatin) to each well and incubate at 37°C for 1 hour, then wash 3 times with PBST, 2 minutes each time; Add 50 μl of 100 nM biotin-labeled CE24 aptamer to the well, incubate at 37°C or room temperature (25°C) for 1 hour, wash with PBST, and wash 3 times for 2 minutes each time. Then add 50 μl strep...
Embodiment 3
[0060] Example 3 Aptamer Direct Method Detection of CE Protein in Intestinal Tuberculosis, Pulmonary Tuberculosis, and Intestinal Crohn's Patient Serum
[0061] Add 100 μl of various sera (including intestinal tuberculosis, pulmonary tuberculosis, intestinal Crohn’s disease and healthy volunteers’ sera) to each well of the 96-well ELISA plate, incubate overnight at 4°C, wash with PBST 3 times, 2 minutes each time . Add 100 μl of 2% BSA to each well, block for 1 hour at 37°C, wash with PBST three times, 2 minutes each time; then add 100 μl of 300 nM biotin-labeled CE24 aptamer to each well, and incubate at 37°C or room temperature (25°C) For 1 hour, wash with PBST 3 times, 2 minutes each time. Then add 50 μl of streptavidin-labeled horseradish peroxidase (HRP) (diluted 1:2000) to act for 30 minutes at 37°C, wash with PBST three times for 2 minutes each time; finally wash with 50 μl of tetramethyl Benzidine (TMB) was used as the chromogenic substrate and incubated at 37°C for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com