Prediction method for protein subcellular site formed based on improved-period pseudo amino acid
A prediction method and pseudo-amino acid technology, applied in the field of bioinformatics, can solve problems such as poor prediction effect and information redundancy, and achieve the effects of difficult to predict offset, reliable prediction method, and protein data balance.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
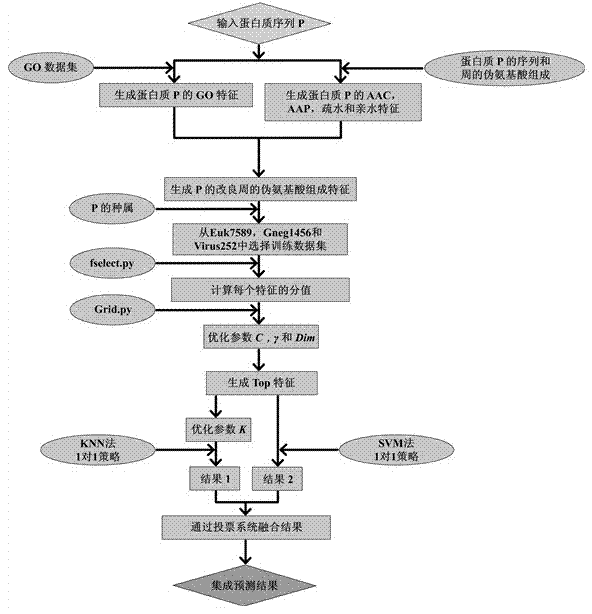
[0032] The construction process of the present invention is described in detail below in conjunction with embodiment:
[0033] 1. Construction of protein datasets: ① eukaryotic protein dataset Euk7579, ② prokaryotic protein dataset Gneg1456, and ③ viral protein dataset Virus252 were respectively obtained through the following addresses:
[0034] ①http: / / web.kuicr.kyoto-u.ac.jp / ~park / Seqdata / ;
[0035] ②http: / / www.csbio.sjtu.edu.cn / bioinf / Gneg-multi / ;
[0036] ③http: / / www.csbio.sjtu.edu.cn / bioinf / virus-multi / .
[0037] Transfer the above-mentioned eukaryotic, prokaryotic, and viral protein data sets into a word file, use the word search and replace functions to delete redundant information, and store the protein number, subcellular site number and amino acid sequence in the newly created file A.xls and A2.xls.
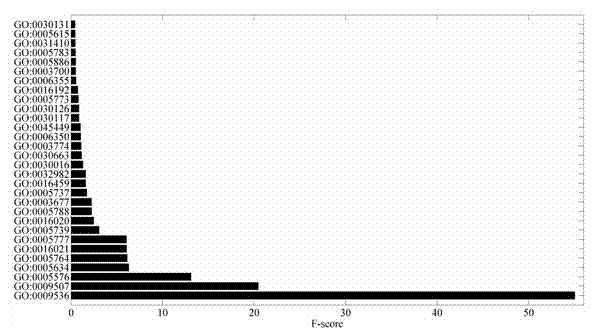
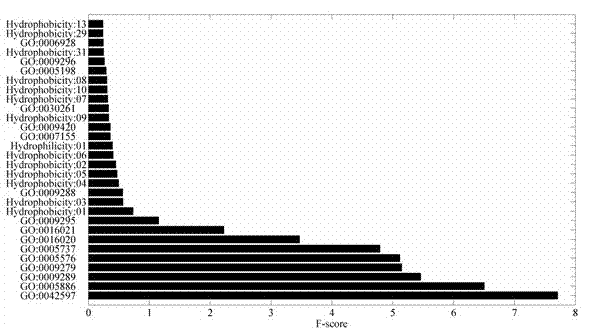
[0038]2. GO feature extraction and vector construction: Download the GO dataset file local_gene_association.goa_uniprot.sorted from ftp: / / ftp.ebi.ac.uk / pub / databases...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com