Method for expressing deoxyribonuclease in plants
A plant, nucleic acid sequence technology, applied in the fields of biochemical equipment and methods, plant peptides, chemical instruments and methods, etc., can solve problems that cannot be easily scaled to commercial levels
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0138] Example 1 - Materials and methods
[0139] Cloning and infiltration
[0140] The nucleic acid constructs comprising the nucleotide sequences encoding the gamma-zein wild-type gene, its fragments and variants are respectively linked to the synthetic sequences encoding the DNase. In the case of using a fragment or variant of γ-zein, the nucleic acid construct also comprises a nucleotide sequence encoding a natural γ-zein signal peptide at the 5′ end, if it is present in the fragment or variant words that exist in the body. For some experiments, a synthetic nucleic acid sequence encoding a linker comprising a protease cleavage site between the gamma-zein coding sequence and the DNase coding sequence was also included in the construct. The coding sequence of the DNase has been optimized for expression in plants. The nucleic acid construct is cloned into a vector at a site where expression of the nucleic acid construct in tobacco plant cells is driven by the minimal 35S p...
Embodiment 2
[0149] Example 2 - Expression Levels of γ-Zein-DNase I Fusion Protein
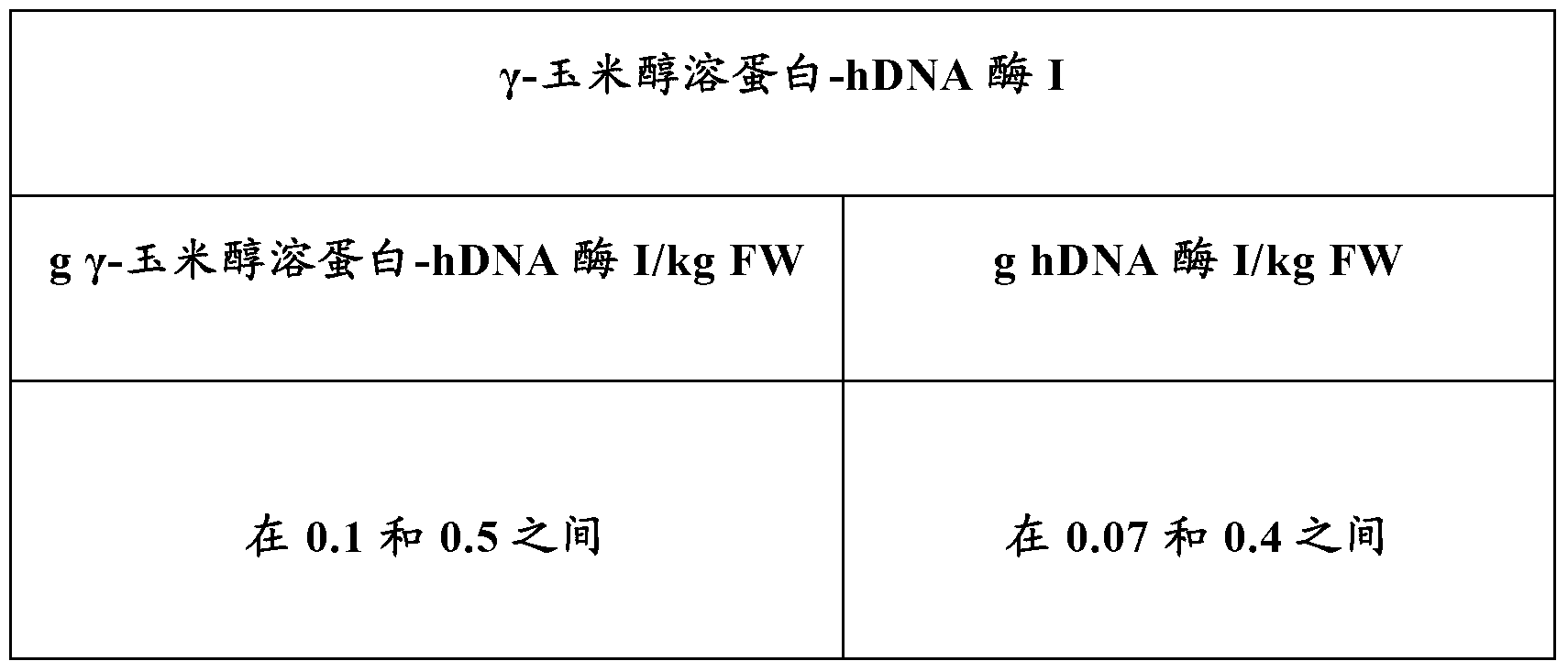
[0150] A gamma-zein-DNase I fusion protein construct (gamma-zein-DNase I) was prepared as described above and transformed into tobacco plants using the Agroinfiltration method. Total protein was extracted and quantified by Western blotting using γ-zein specific antibodies. A control experiment (hDNase I) was also performed using DNase I expressed under the same conditions without the gamma-zein tag. The expression levels from the mean values of Agroinfiltration events were as follows:
[0151]
[0152] FW = net weight (free weight). Based on these results, it was concluded that the expression of γ-zein-hDNase I was higher than that of human DNase I in the absence of γ-zein.
Embodiment 3
[0153] Example 3 - Analysis of different non-naturally occurring repeat sequence motifs in gamma zein
[0154] γ-Zein-DNase I fusion constructs were made using different non-naturally occurring repeat sequence motifs in γ-zein. The following constructs were used: γ-zein peptide only (zein-wild type); γ-zein-(PPPVAL)n; γ-zein-(PPPVEL)n; γ-maize prolamin-(PPPAPA)n; and gamma-zein-free.
[0155] The constructs were transformed separately into different tobacco plants using the Agrobacterium infiltration method. Three independent transformations were performed. Total protein was extracted and quantified by Western blotting using γ-zein specific antibodies. Expression levels from the mean of 3 Agroinfiltration events were as follows:
[0156] construct
[0157] Results are expressed as relative quantification compared to γ-zein wild type. Unexpectedly, human DNase I was expressed at the highest level in the absence of γ-zein. Other non-naturally occurring repeat mot...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com