Haemophilus parasuis with double-gene deletion and construction method thereof
A technology of Haemophilus suis and gene deletion, applied in the field of Haemophilus parasuis and its construction, which can solve the problems of unclear pathogenic mechanism of Hps, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] A Haemophilus parasuis with double gene deletion is constructed by the following method:
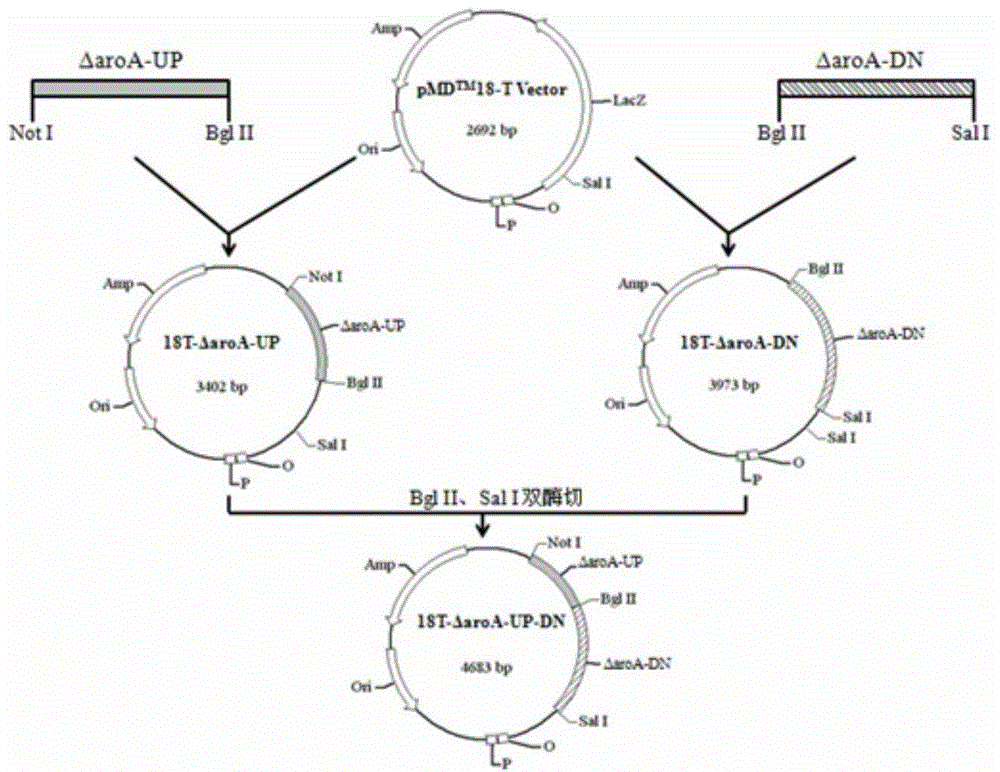
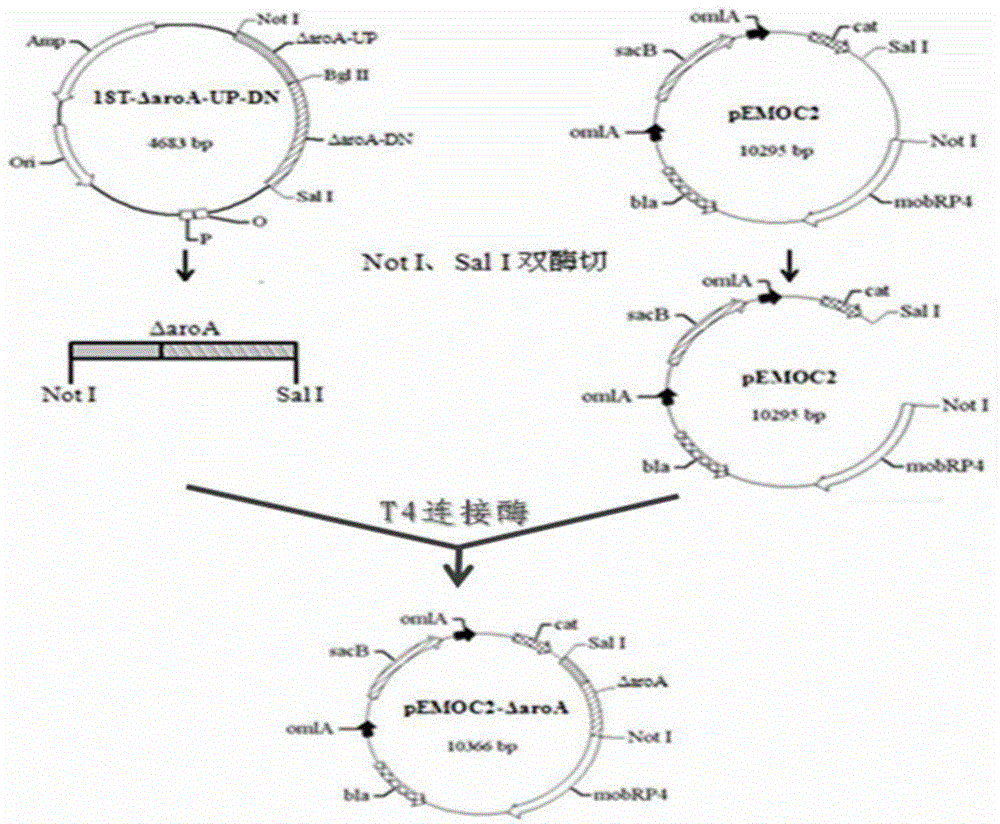
[0059]1) PCR identification of strain H10: referring to the method of PCR identification of Hps established by Oliveira, S. et al. in 2001, a pair of primers SEQ ID NO: 11 and SEQ ID NO: 12 were synthesized, and 16S was amplified using the overnight cultured bacterial solution as a template For rRNA fragments, the PCR reaction system is 25 μL: 10xTaqbuffer 2.5 μL, 2 mmol / L dNTPs 0.5 μL, 16S rRNA-1 and 16S rRNA-2 (10 μmol / L) 1 μL each, Taqase 0.5 μL, template 1 μL, ddH2O to 25 μL. The PCR reaction program is: 94°C for 4min, then enter the PCR cycle: 94°C for 30s, 58°C for 30s, 72°C for 1min, 30 cycles; finally, extend at 72°C for 10min; the amplified product is identified by 0.8% agarose gel electrophoresis, and the UV Observed under the lamp; a single band was amplified with a size of about 800bp, which was consistent with the expected 821bp fragment, proving that the strain H10 w...
Embodiment 2
[0077] The steps are the same as in the example, except that the identification of the clones in step 7) adopts the conventional method. Pick the single colonies grown after transformation, inoculate them in 3ml of liquid medium containing the corresponding antibiotics, and culture them at 37°C and 250r / min for 6h Left and right until the medium becomes turbid; take 100 μL of the bacterial solution, centrifuge at 5000r / min for 10min, discard the supernatant, resuspend the bacterial cells with 30μL of sterilized double distilled water, boil the cell suspension for 10min, then ice-bath for 10min, 12000r / min Centrifuge for 1 min, and take the supernatant as a PCR template for PCR identification.
[0078] Result identification
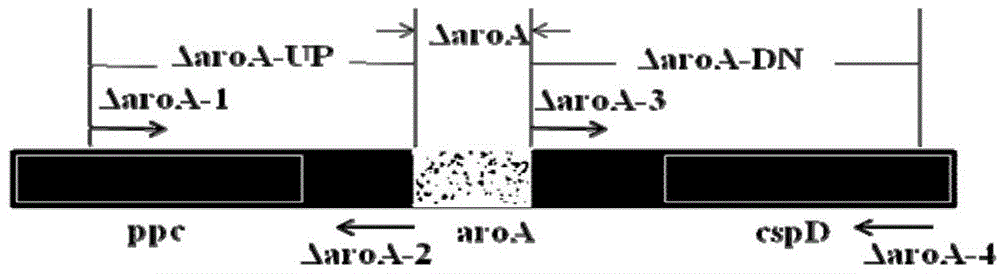
[0079] 1. Amplification of the front and rear arm fragments of the aroA gene: such as Image 6 As shown, the electrophoresis results showed a band of about 700bp, consistent with the expected 710bp fragment. The rear arm ΔaroA-DN of the aroA gene was amp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com