One group of nucleic acid aptamers capable of specifically identifying Beijing genotypes tuberculosis bacterial strain antigen and application of nucleic acid aptamers
A Beijing genotype and nucleic acid aptamer technology, applied in the biological field, can solve problems such as unusable monitoring, difficult popularization, and high cost, and achieve the effects of overcoming low sensitivity, simple implementation, and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
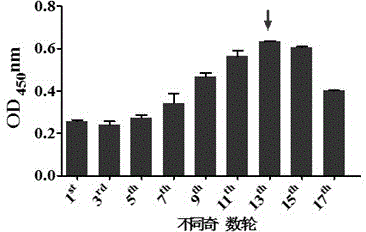
[0039] [Example 1] Screening of aptamers
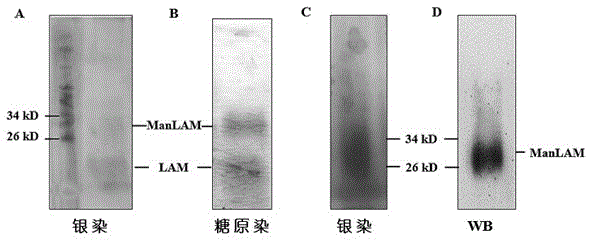
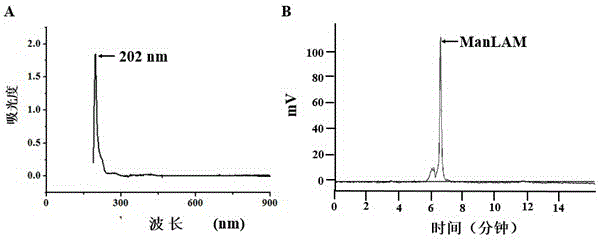
[0040] 1. Extraction, purification and identification of the surface lipid sugar ManLAM of Mycobacterium tuberculosis Beijing genotype strain
[0041] (1) Culture in 7H9 liquid medium (Weigh 4.79 grams of Mycobacterium tuberculosis medium Middlebrook 7H9 and dissolve in 900ml of deionized water, add 2ml of glycerin and mix well, sterilize at 121°C for 10 minutes, add in a volume ratio of 9:1 Bacteria enrichment solution (Middlebrook OADC, spare) to OD about 1.8, inactivated in a 65°C water bath for 1 hour, then centrifuged at 10,000 rpm for 10 minutes to collect Mycobacterium tuberculosis Beijing genotype bacteria, added a small amount of PBS to wash twice, and freeze-dried;
[0042] (2) Resuspend the dried H37Rv in 10 times the volume of chloroform / methanol (v / v=2:1), and degrease in a water bath at 37°C for 12 hours;
[0043] (3) Collect the precipitate by centrifugation at 10,000 rpm for 10 minutes, resuspend the precipitate in ch...
Embodiment 2
[0126] [Example 2] ELONA (adapter) method detection specificity
[0127] 1. Coated with Beijing genotype Mycobacterium tuberculosis, Mycobacterium smegmatis, BCG, Mycobacterium avium, Escherichia coli DH5α, Staphylococcus aureus, Bacillus denatus, Enterococcus, Klebsiella pneumoniae, Staphylococcus epidermidis, Pseudomonas aeruginosa, Streptococcus, Neisseria, H37Rv Mycobacterium tuberculosis (10 6 cfu), were respectively coated on 96-well plates, placed in a 37°C wet box for coating for 1 hour, and the microwells were washed 3 times with PBST;
[0128] 2. Block the microwells with 1% BSA (200 μl / well) at 37°C for 1 hour, and wash the microwells 3 times with PBST;
[0129] 3. After measuring the concentration of the above five aptamers of biotin-labeled ManLAM obtained by asymmetric PCR, add 30 pmol, incubate at 37°C for 1 hour, and wash the microwells 3 times with PBST;
[0130] 4. Add HRP-labeled streptavidin (diluted 1:2000), 100 μl / well, act at 37°C for 45 minutes, wash ...
Embodiment 3
[0134] [Example 3] Aptamer ELONA (aptamer) method detects the sensitivity of ManLAM lipid sugar
[0135] 1. Coating with different concentrations of ManLAM lipid sugar (100 μg / mL, 50 μg / mL, 25 μg / mL, 12.5 μg / mL, 6.25 μg / mL, 3 μg / mL, 1.5 μg / mL, 0.75 μg / mL, 0.37 μg / mL mL, 0.18 μg / mL) in a 96-well microplate, coated at 37°C for 1 hour, and washed with PBST for 3 times;
[0136] 2. Block the microwells with 1% BSA (200 μl / well) at 37°C for 1 hour, and wash the microwells 3 times with PBST;
[0137] 3. After measuring the concentration of the five biotin-labeled aptamers obtained by asymmetric PCR, add 30 pmol, incubate at 37°C for 1 hour, and wash the microwells 3 times with PBST;
[0138] 4. Add HRP-labeled streptavidin (diluted 1:2000), 100 μl / well, act at 37°C for 45 minutes, wash the microwells 3 times with PBST;
[0139] 5. Add TMB substrate, react at 37°C for 1min to 3min, and observe the color change. Then stop with 2M concentrated sulfuric acid;
[0140] 6. Use a micro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com