Grouping for classifying gastric cancer
A grouping method, gastric cancer technology, applied in biochemical equipment and methods, medical preparations containing active ingredients, analytical materials, etc., can solve the problem of not being able to capture the complete diversity of gastric cancer subtypes, and the difficulty of deriving immortal cell lines from gastric tumors And other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
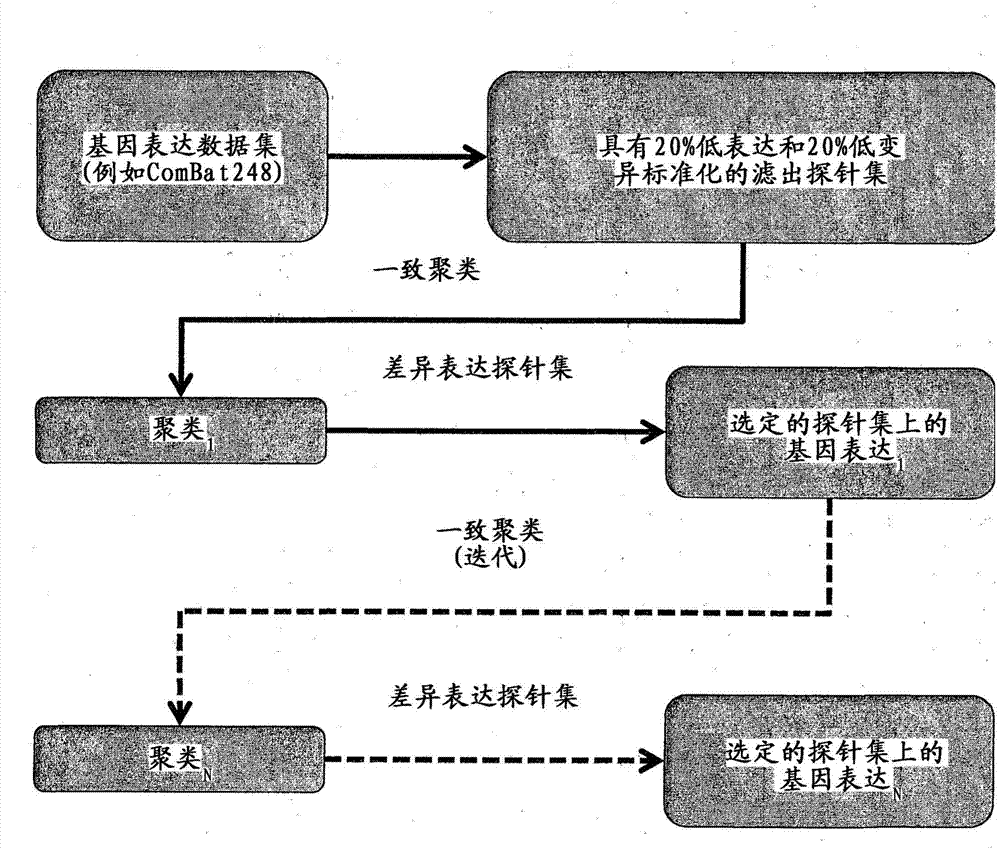
[0208] Determining the number of gastric cancer subtypes
[0209] Fifty-six gastric adenocarcinomas were analyzed and combined with previously reported expression profiles to form a collection of 248 profiles (Gene Expression Omnibus accession numbers GSE15459 and GSE22183). All samples were analyzed on the Affymetrix U133 Plus 2.0 expression array.
[0210] Gene expression microarray data obtained from multiple sources have significant batch effects. Therefore, to ensure that batch effects or non-biological variation between sample groups arising from subtle but systematic differences in experimental manipulations do not obscure biological structures or lead to artifacts, the data were processed through ComBat to eliminate batch effect. The nascent dataset is named "SG248".
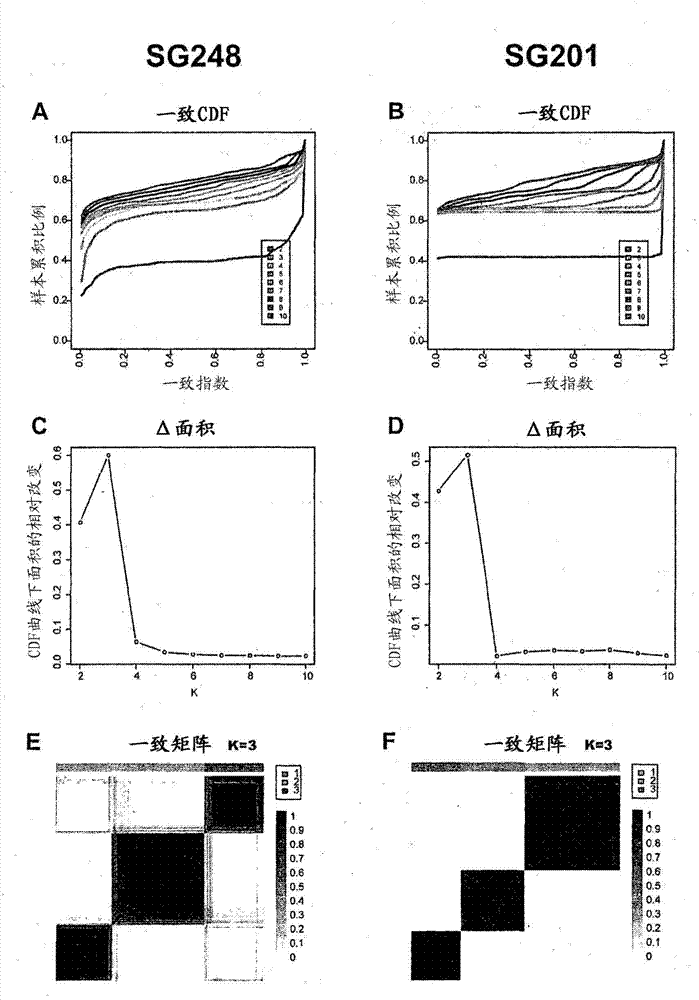
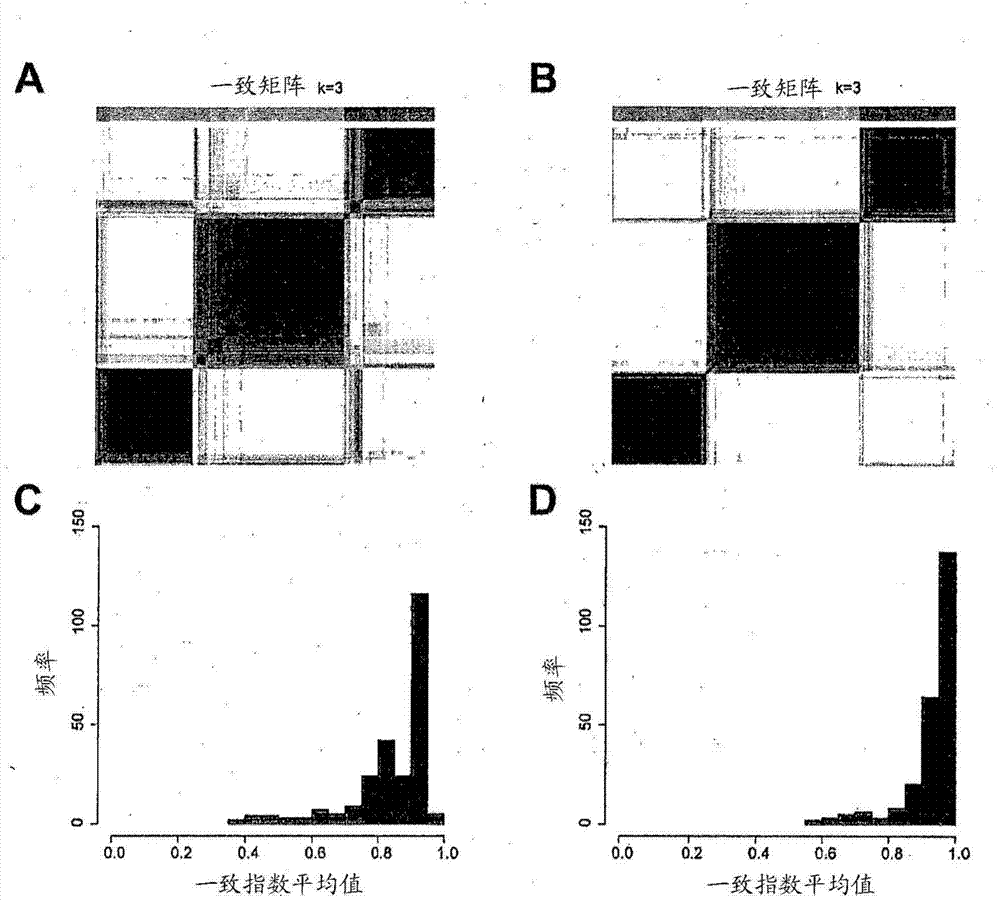
[0211] Assessment of whether biological substructures were retained after ComBat treatment was done as follows. First, consensus clustering was applied to each of the two individual batches (Singap...
Embodiment 2
[0230] Genomic Copy Number Alterations (CNAs)
[0231] 138 of the SG201 tumor samples were analyzed using the Affymetrix SNP6 DNA microarray (Affymetrix, 2009), and 98 non-malignant samples were used as reference. Using these data, the number of chromosomal segments affected by CNAs was determined for each tumor.
[0232] Preprocessing and normalization were performed using Affymetrix Genotyping Console 3.0 (Affymertrix, 2008). For each tumor sample, "Log2Ratios" (term in Affymetrix Genotyping Console) were generated for SNPs (single nucleotide polymorphisms) and copy number probe sets on the array. Cyclic binary partitioning was applied to Log2Ratios using the R package DNAcopy (http: / / www.bioconductor.org / packages / 2.3 / bioc / html / DNAcopy.html).
[0233] The segmented data were then mapped to chromosome segments using the hg18 chromosome segment locations from the UCSC Genome Browse database. The copy number of each chromosome segment was estimated as the length-weighted a...
Embodiment 3
[0243] methylation profile
[0244] The DNA methylation profiles of 139 of the SG201 tumor samples were determined. The Illumina Infinium HumanMethylation 27 array (Weisenberger et al., 2008) was used to evaluate CpG methylation levels across 26,486 orthosomes across the genome, including 94 non-malignant samples from the Singapore cohort as a reference.
[0245] The extent to which the DNA methylation profiles of each subtype differed from those of 94 non-malignant gastric tissue samples was investigated. To do this, the methylation levels of each CpG in all tumors in each subtype were compared to the methylation levels of non-malignant samples. The DNA methylation level (β value) of each probe was calculated using Illumina's Genome Studio software. Differential methylation analysis relative to 94 non-malignant samples was performed on the aggressive subtype, proliferative subtype, and metabolic subtype sample sets, respectively. CpGs with significantly different methyla...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap