High-throughput sequencing-based methods and kits for labeling and capturing one or more specific genes of multiple samples
A high-throughput, kit-based technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of high sequencing cost and low throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
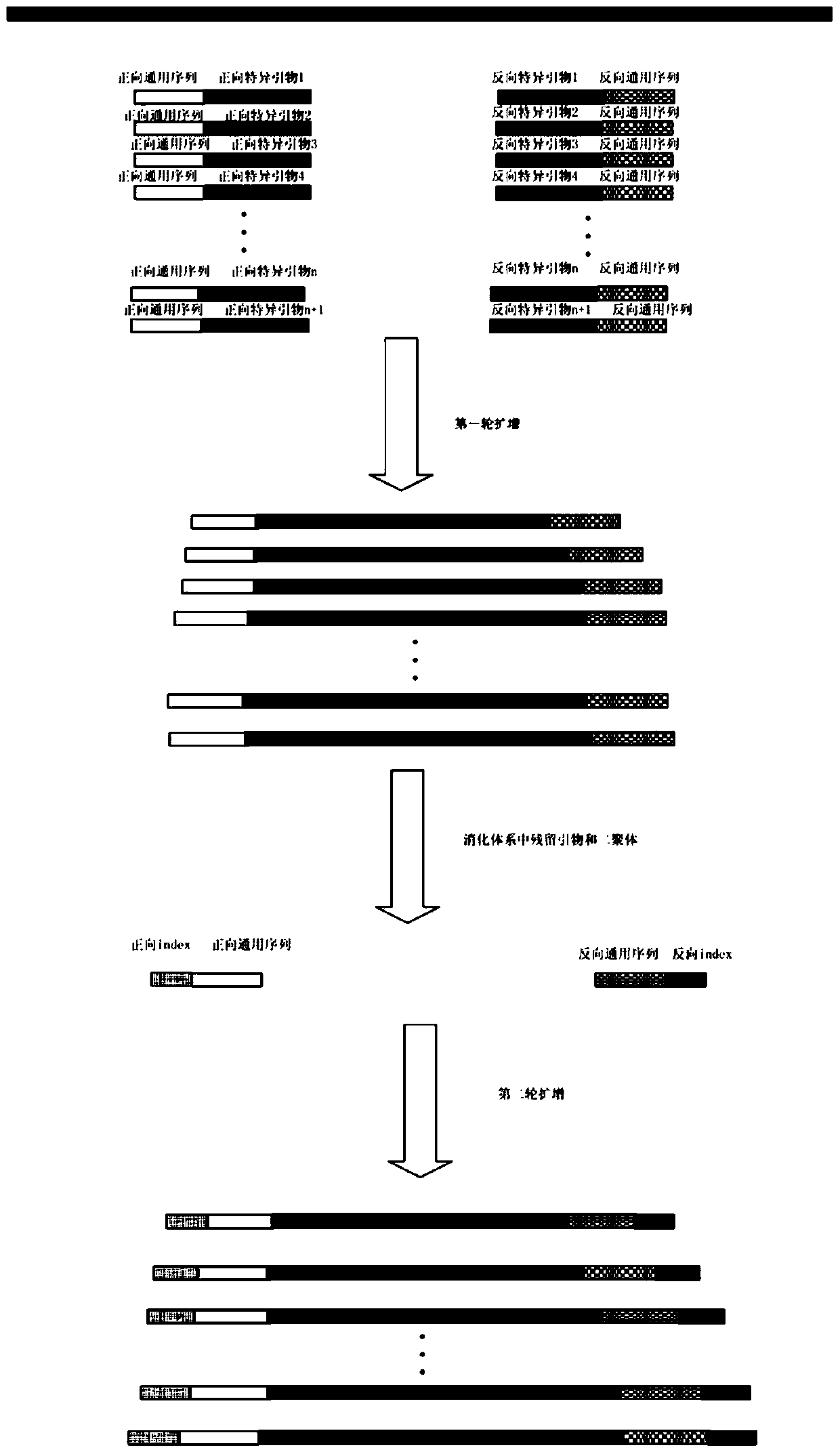
[0065] 27 exons of pheochromocytoma-related SDHB, SDHC, SDHD, RET, and VHL 5 genes were sequenced, with a total of 25 clinical samples:
[0066] 1) Primer design:
[0067] Corresponding amplification primers were designed for 27 exons of the 5 genes, and the related parameters were: Tm value 55.0°C-60.0°C, GC value 40.0%-60.0%, primer size 20±3bp. A general sequence is added to the 5' ends of the forward amplification primer and the reverse amplification primer respectively. The designed primers are as follows, and the underlined is the general sequence introduced:
[0068]
[0069]
[0070] Design the tag sequence according to the following rules: the number of forward tag primers × the number of reverse tag primers ≥ the number of samples, in this example, design 5 pairs of tag sequences based on 25 samples, and the 3' end plus the universal sequence to form the second The index primers used in the round of amplification are amplified through the combination of 5 pair...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com