Massive Feature Screening Method for Ligand Molecules in Drug Design
A feature screening and ligand molecule technology, applied in the field of computer-aided drug design, can solve the problems of high time consumption and achieve the effect of increasing comprehensibility and improving learning efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0018] The present invention will be further described in detail below in conjunction with the accompanying drawings of the specification.
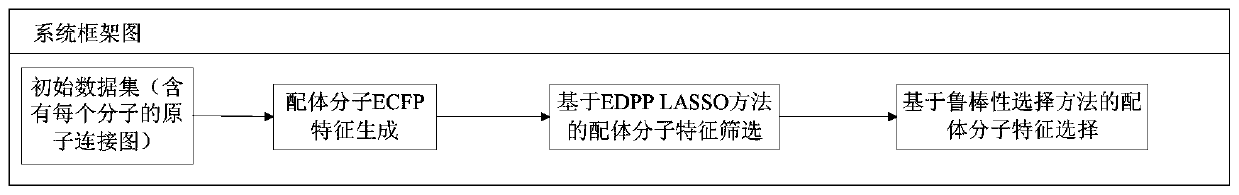
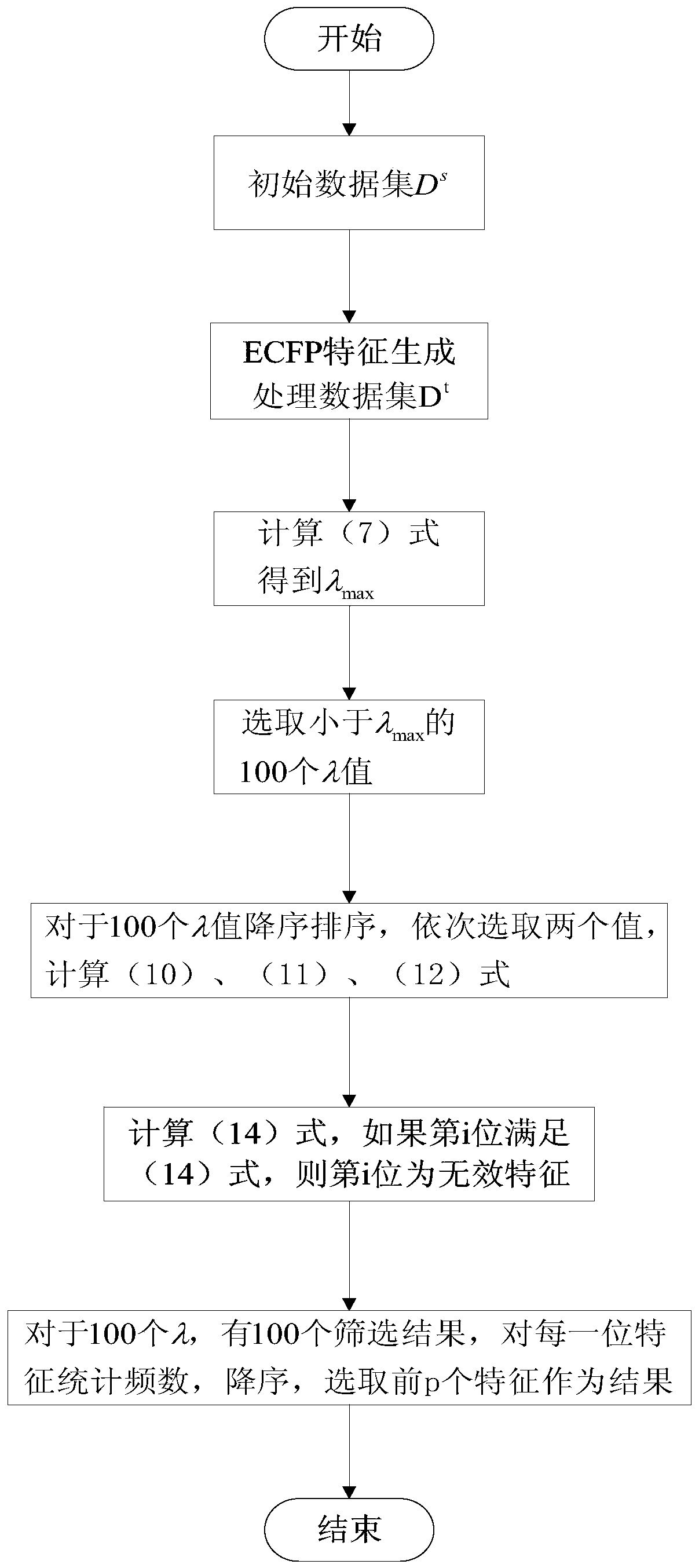
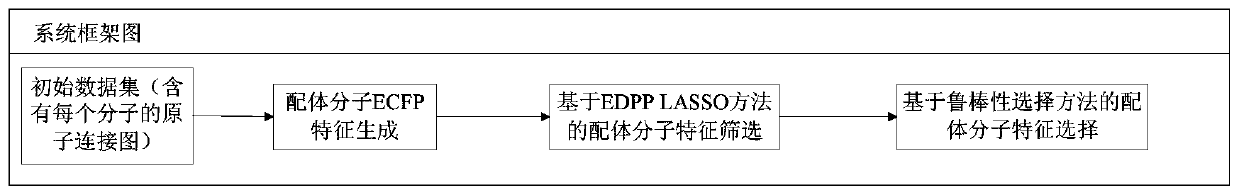
[0019] figure 1 It is a framework diagram of the system of the present invention. Based on this framework, the present invention provides a method for screening massive features of LASSO ligands based on EDPP criteria. The specific implementation steps of the method include the following:
[0020] Step 1: Generation of ECFP feature of ligand molecule. Given initial data set among them Is the atom connection diagram of each molecule, Y i Is the mark of each sample. Process the initial data set to get the ECFP feature describing the sample, that is, data set D t ={(X i ,Y i )|X i ∈R 1*m ,1≤i≤n}.
[0021] Step 2: Screening of ligand molecular characteristics based on EDPP LASSO method. For data set D t , Applying the EDPP criterion, for satisfying the condition (λ∈(0,λ 0 ]) of λ={λ i |0≤i i > λ i+1 }, get the feature screening result of each λ ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com