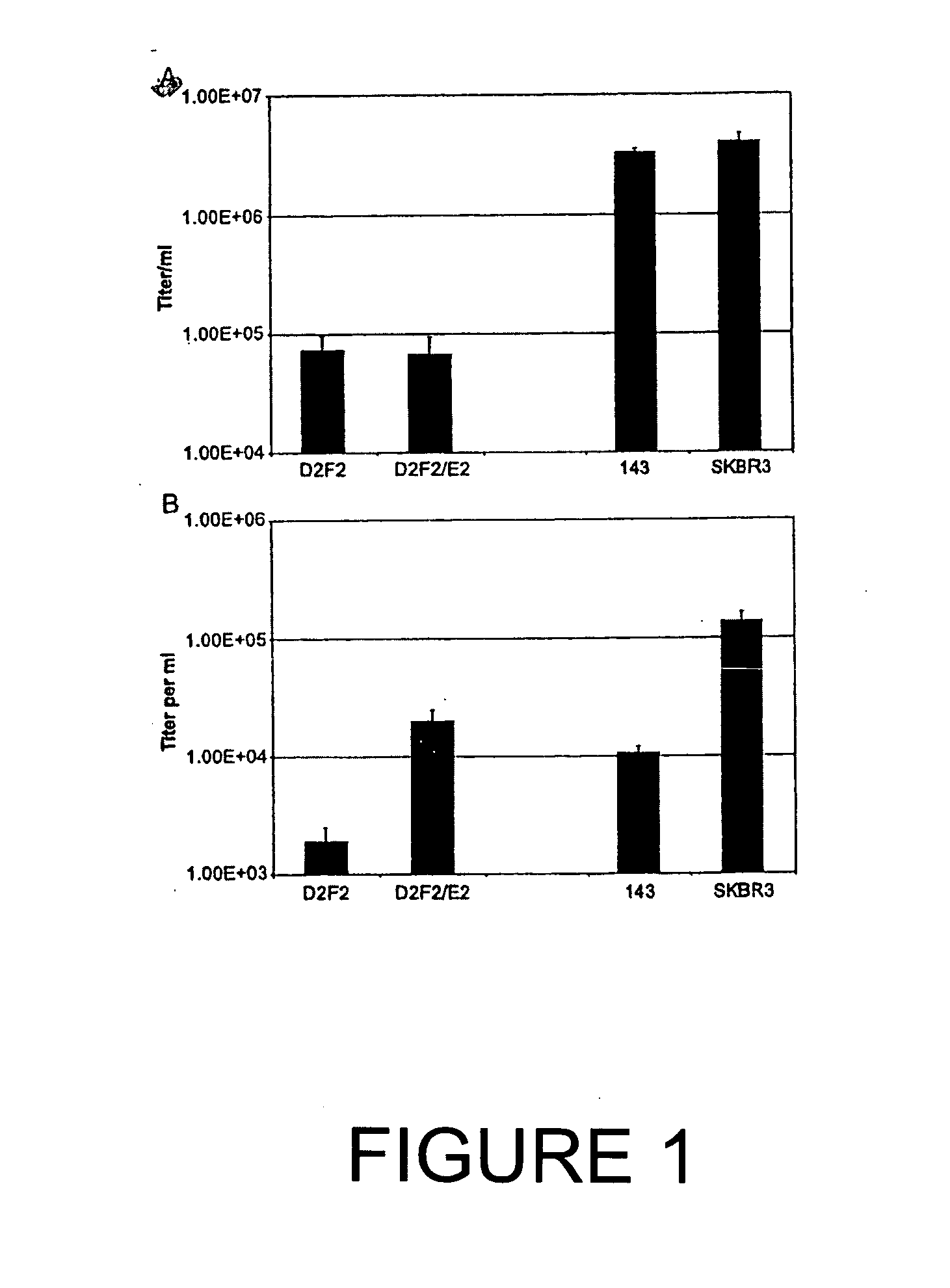
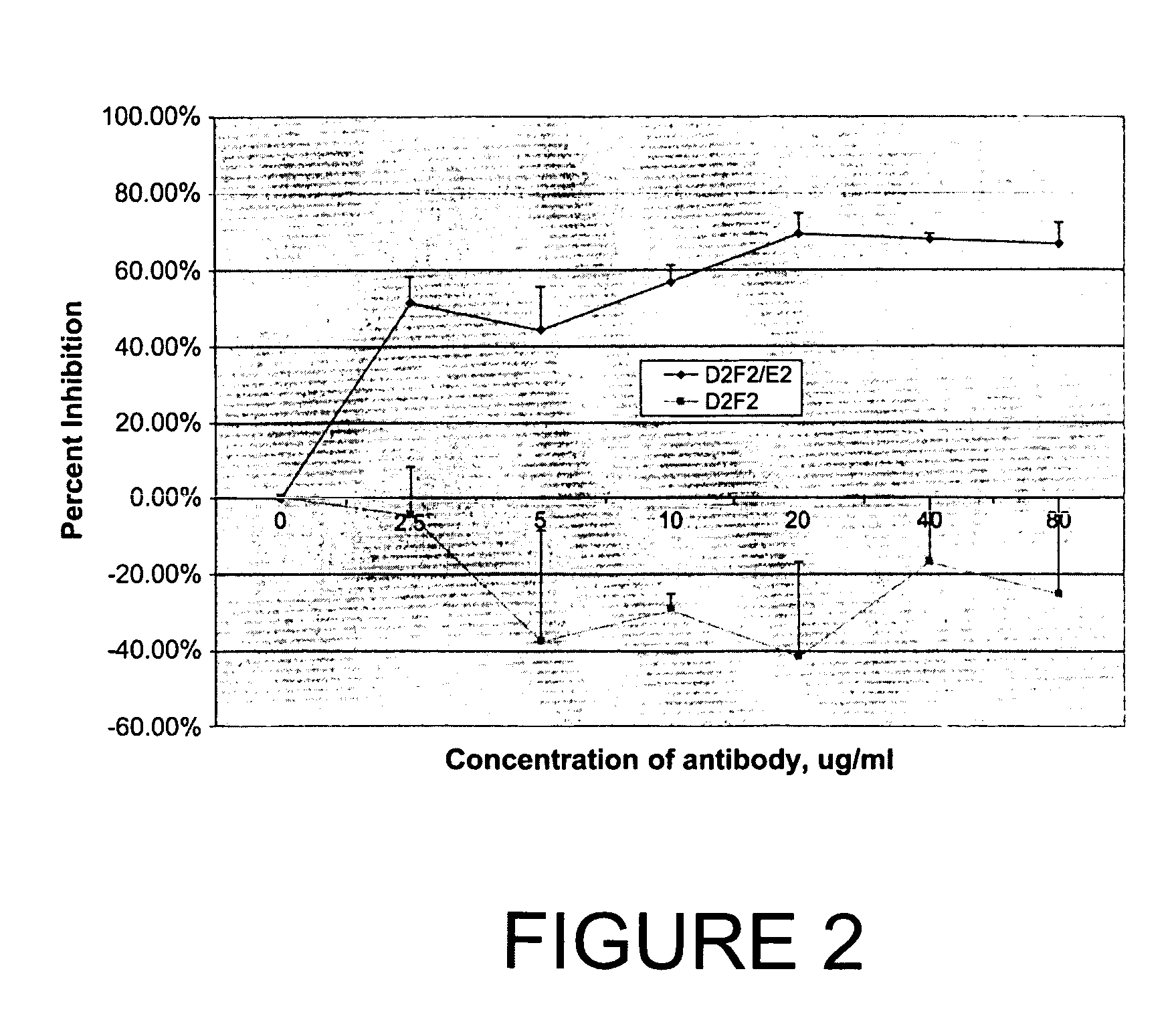
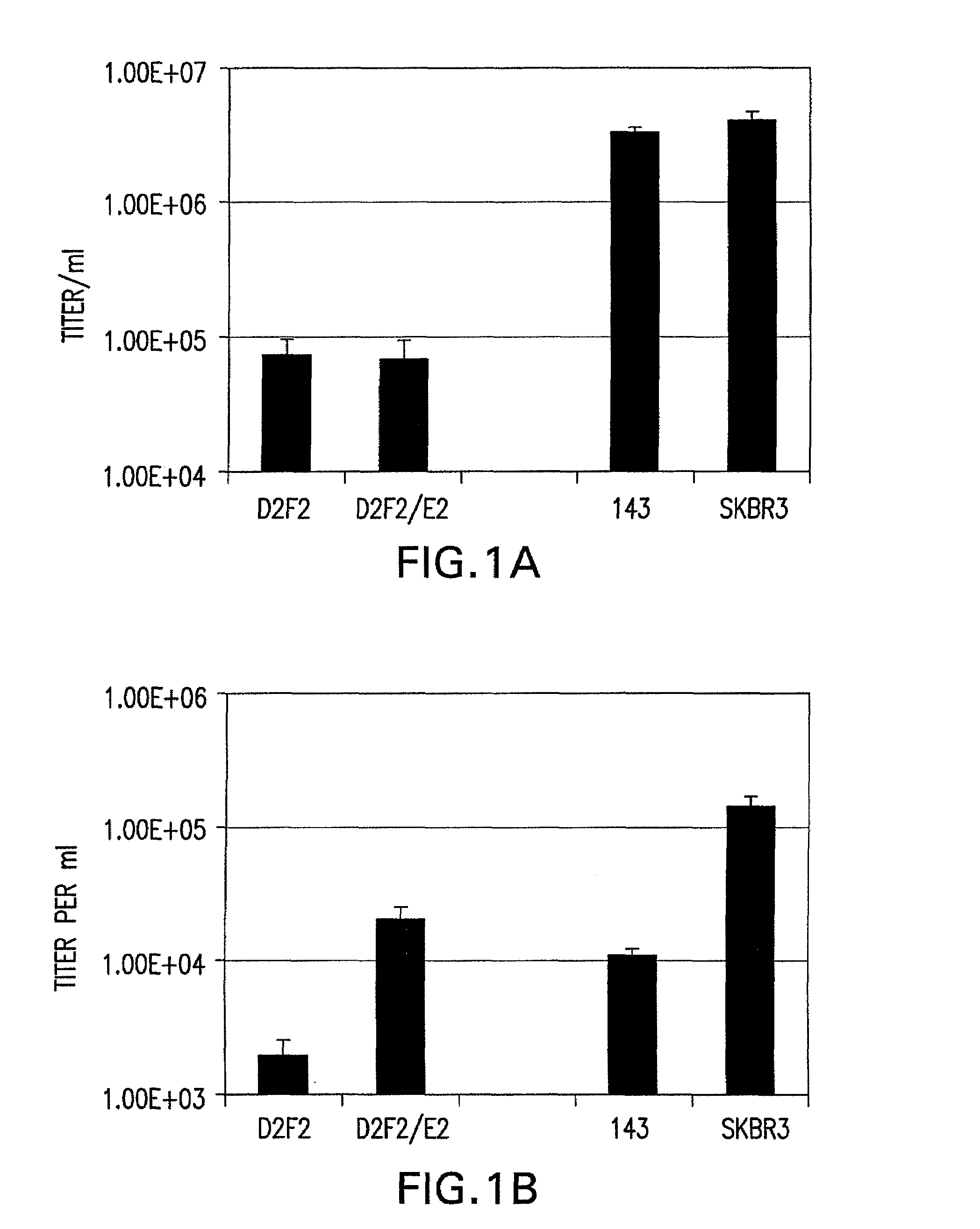
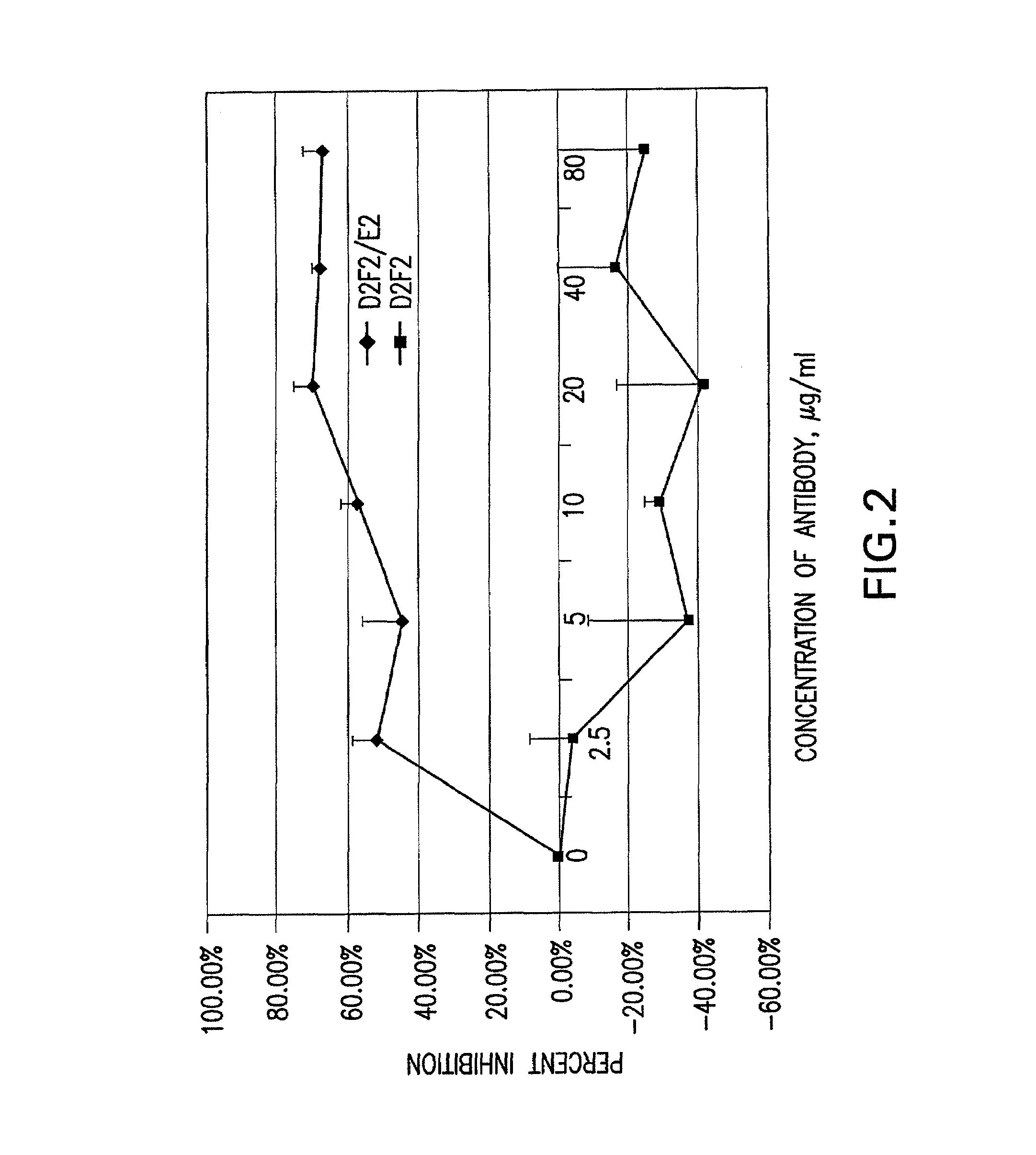
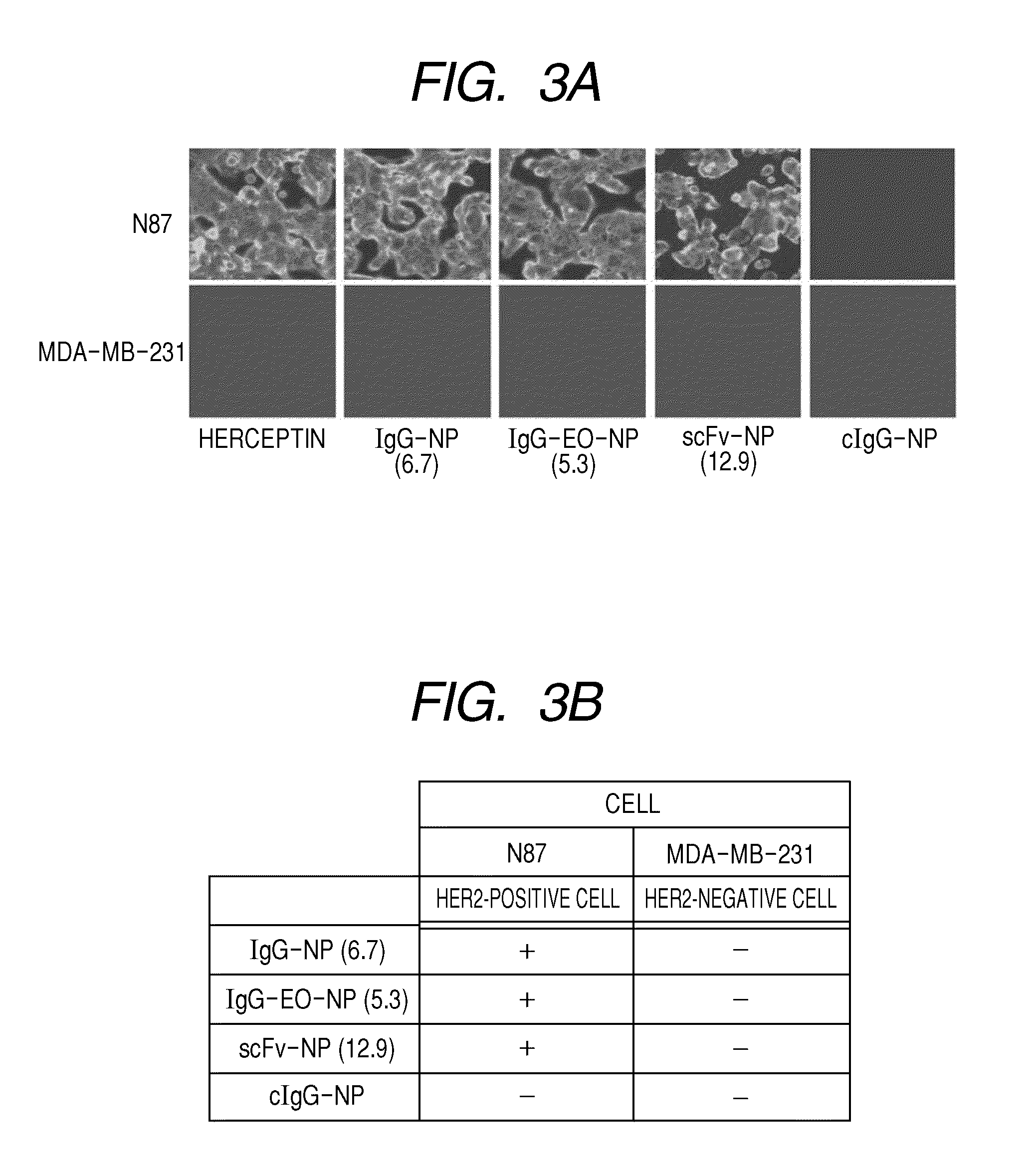
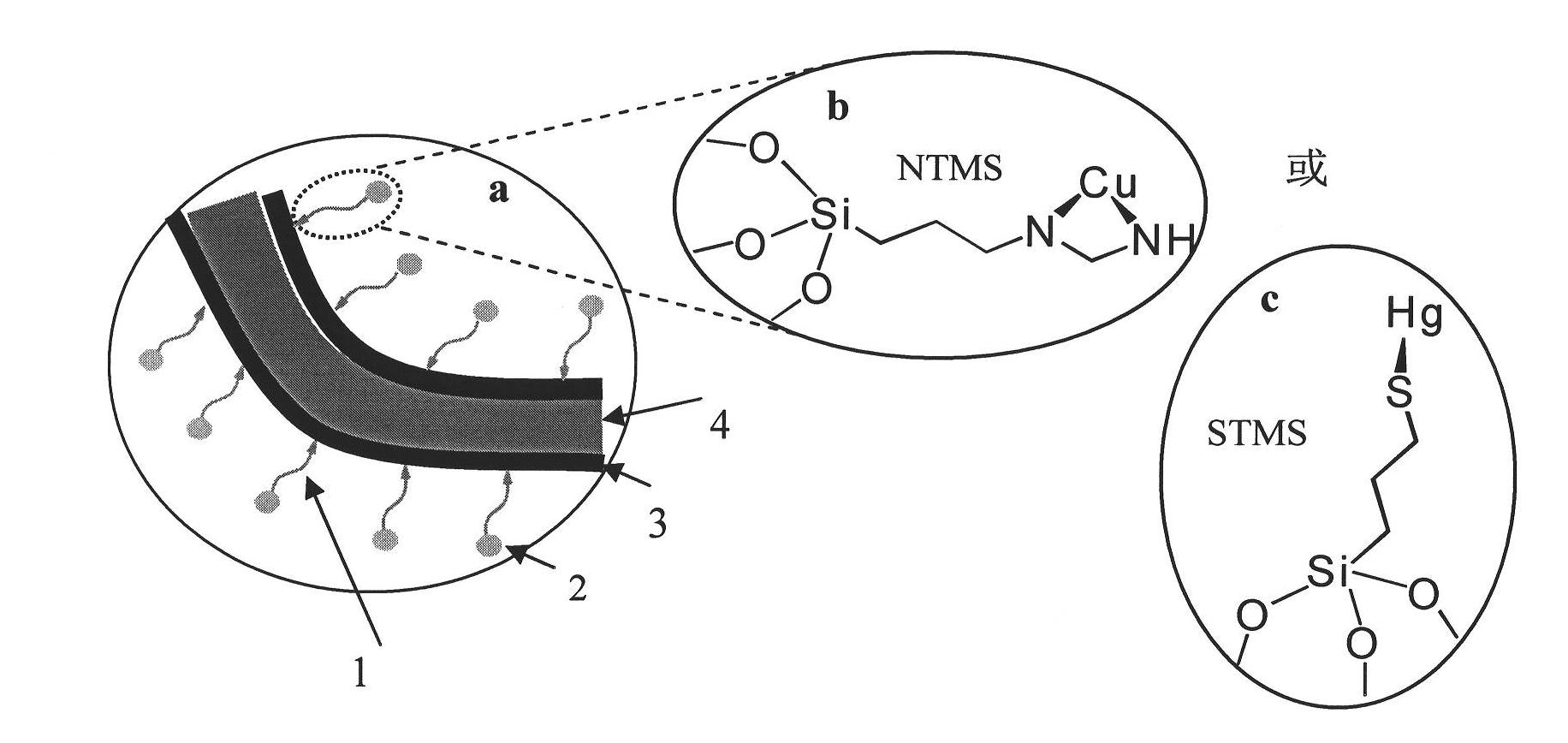
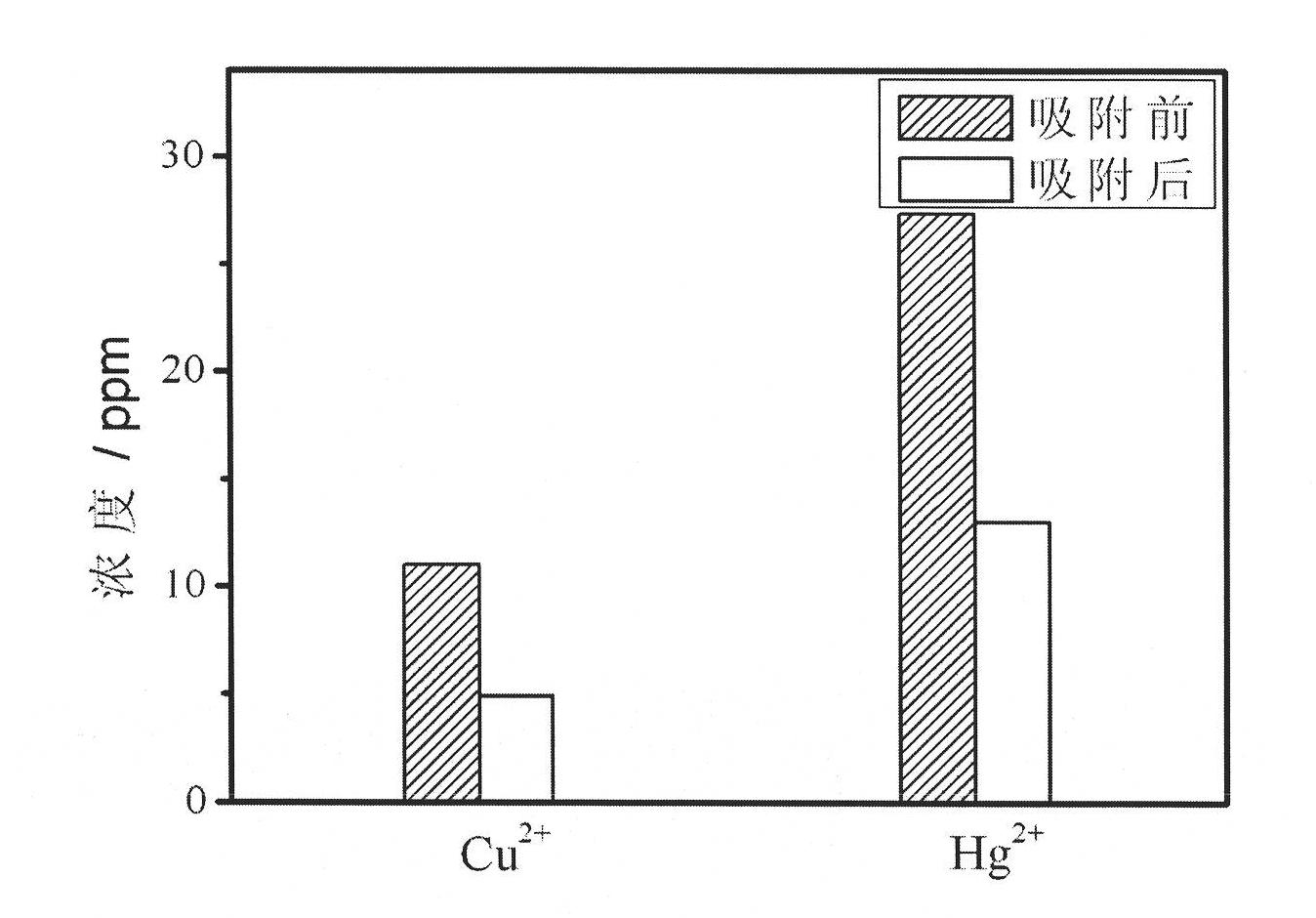
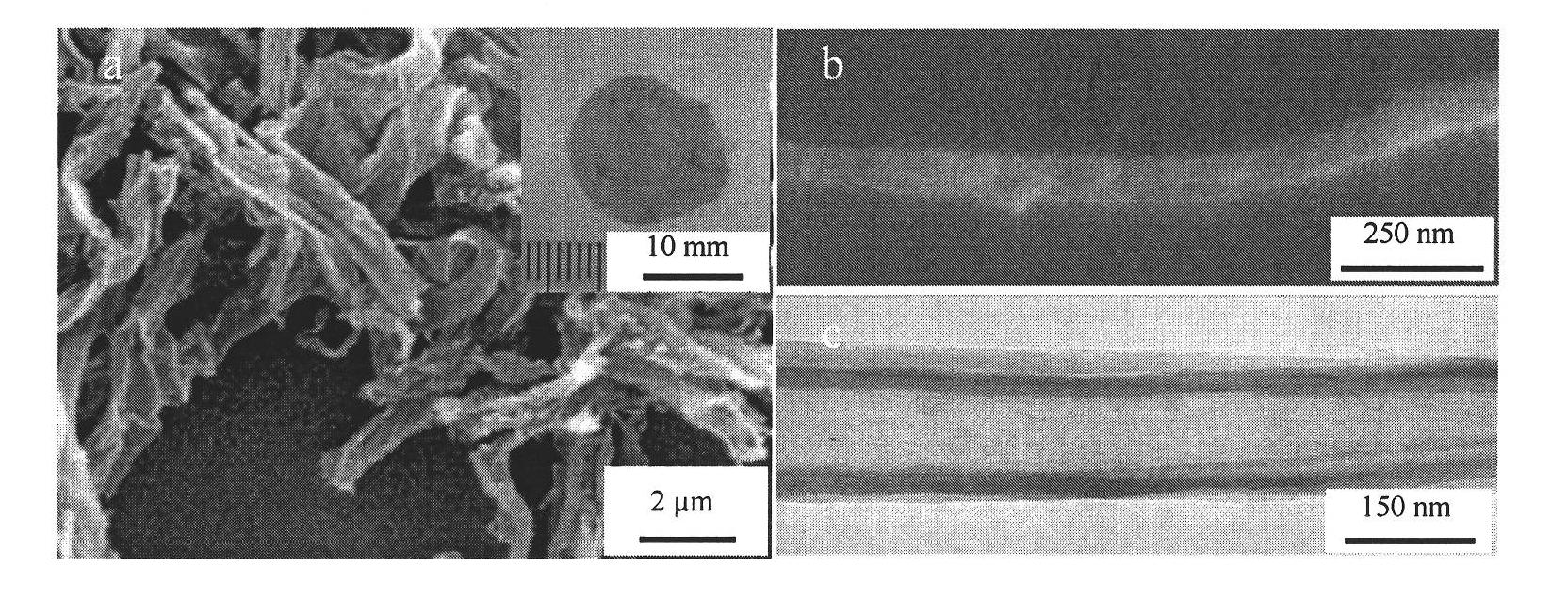
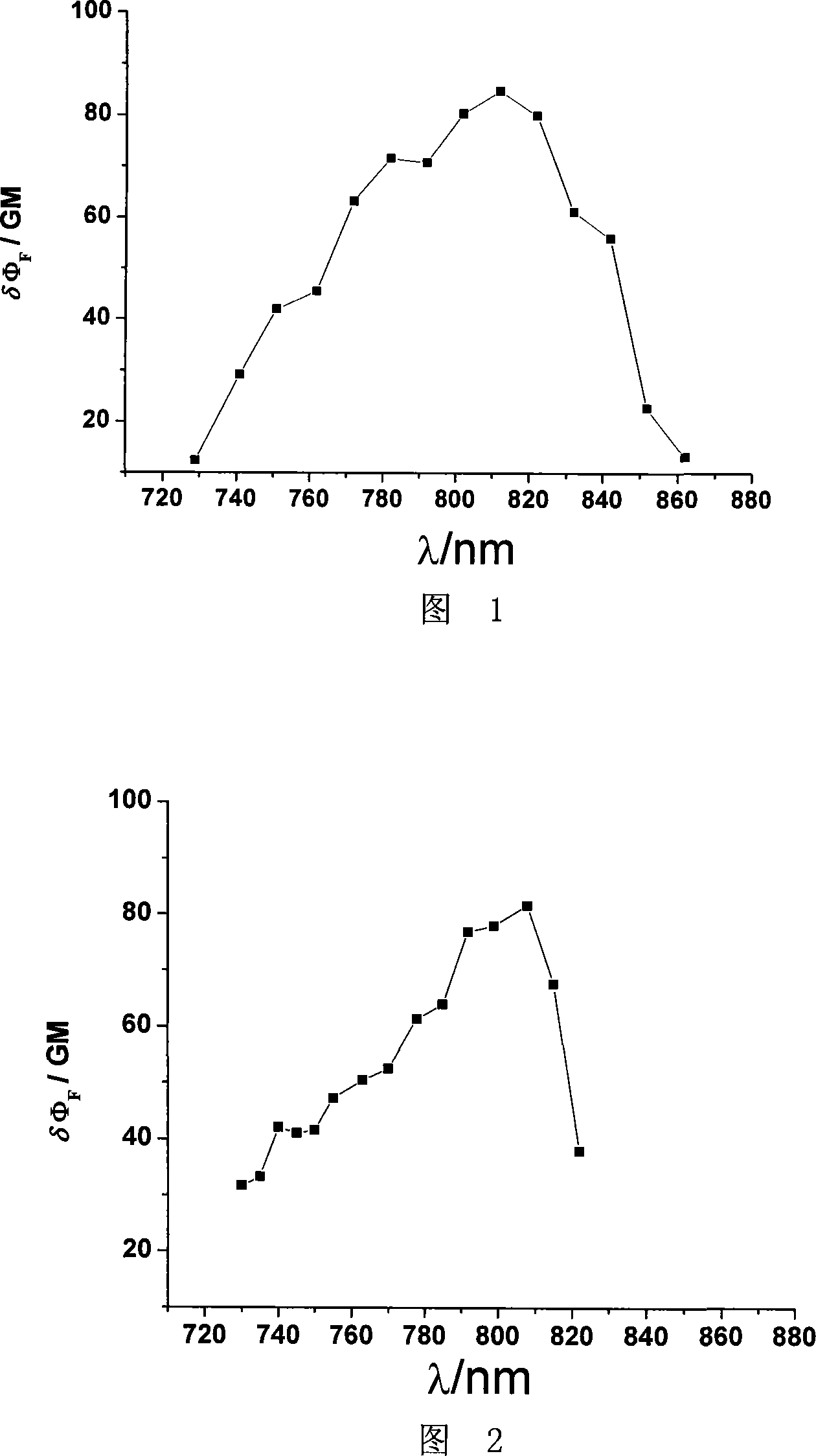
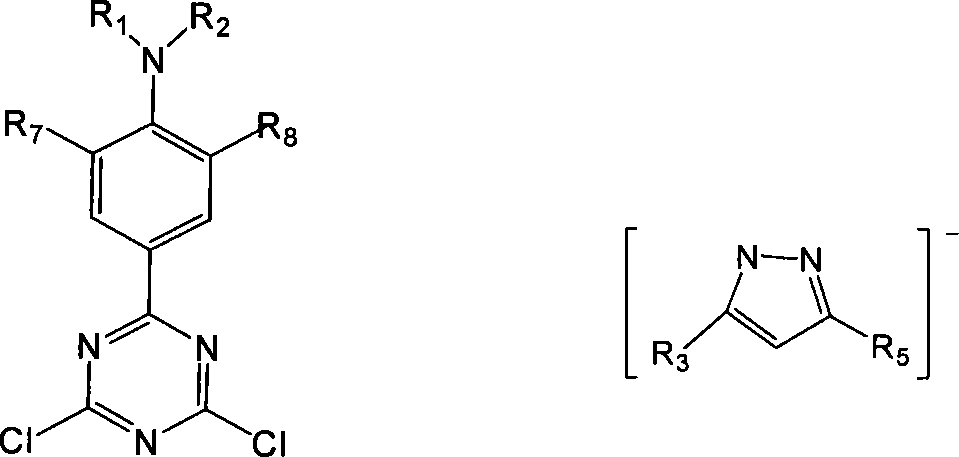
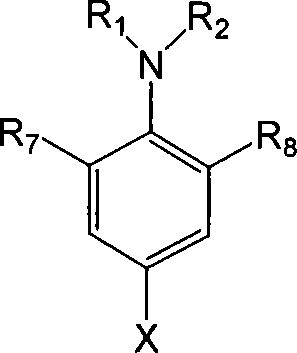
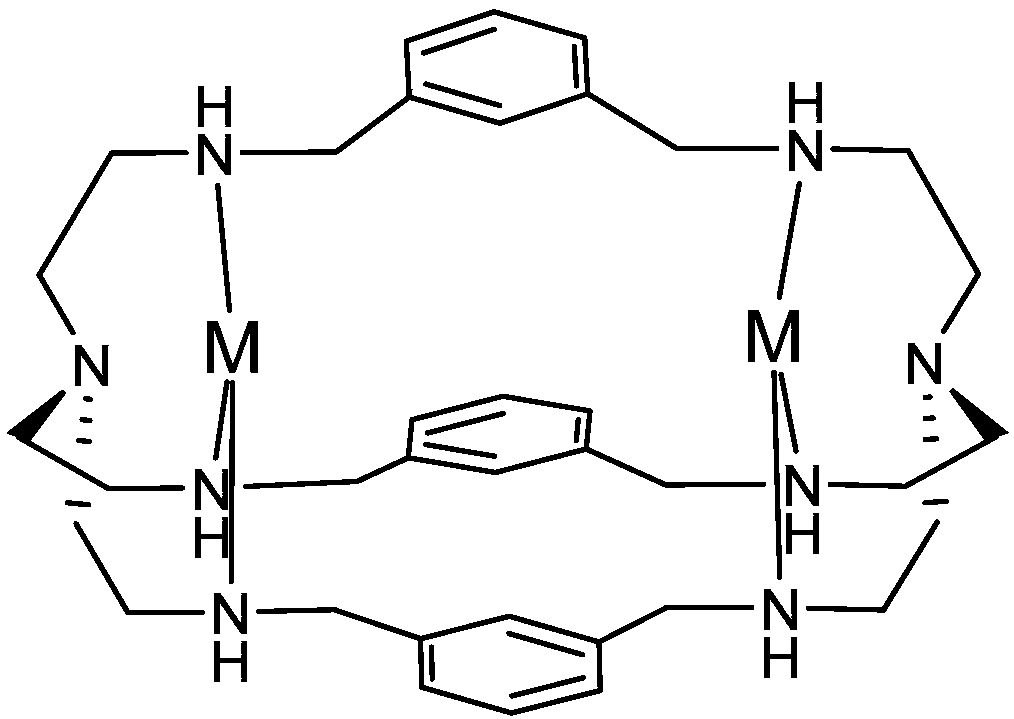
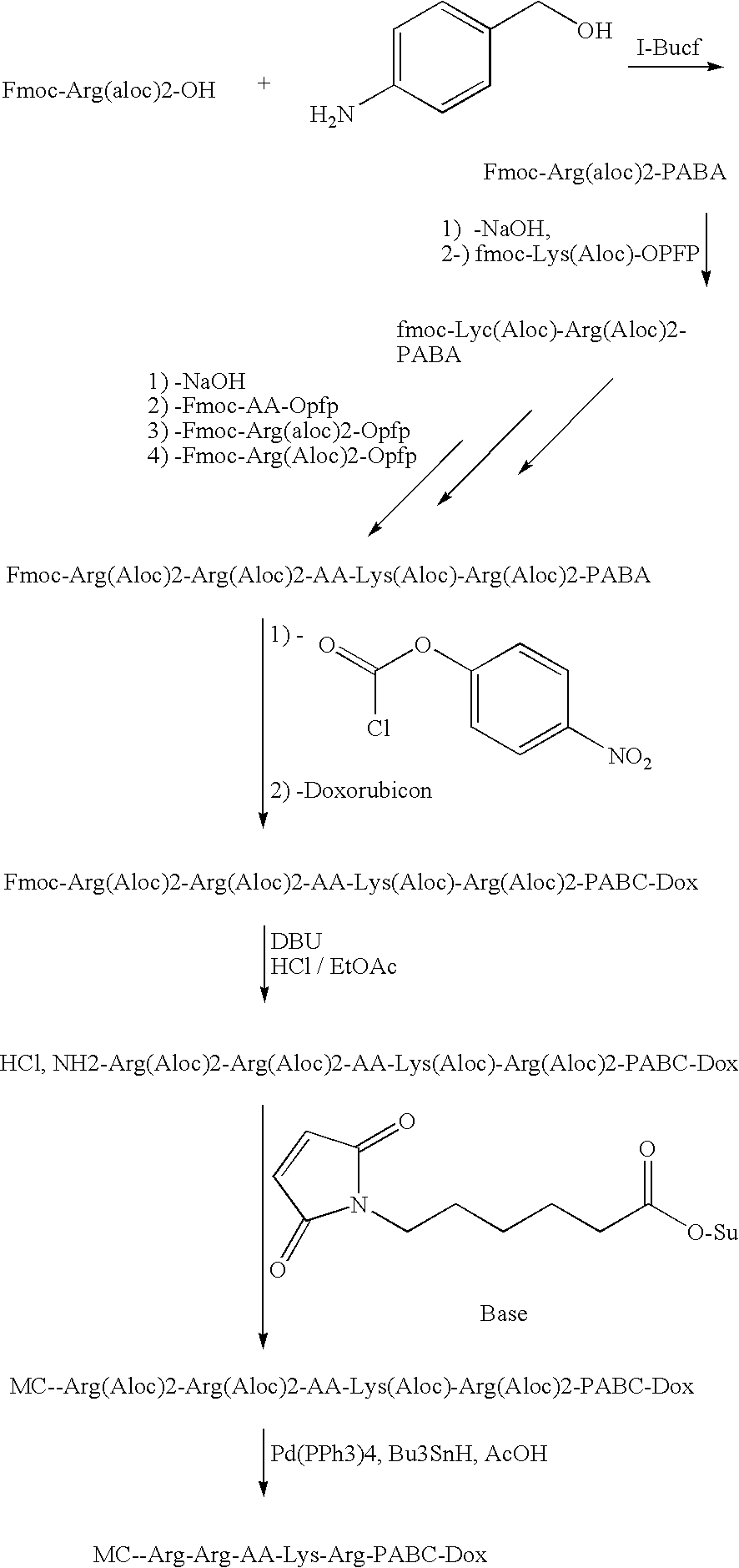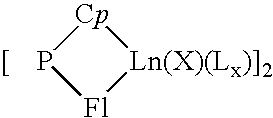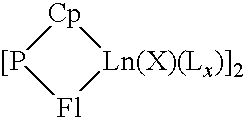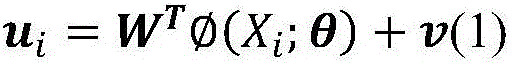Patents
Literature
302 results about "Ligand molecule" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
In biochemistry, a ligand is any molecule or atom which binds reversibly to a protein. A ligand can be an individual atom or ion.
Isolation of proteins
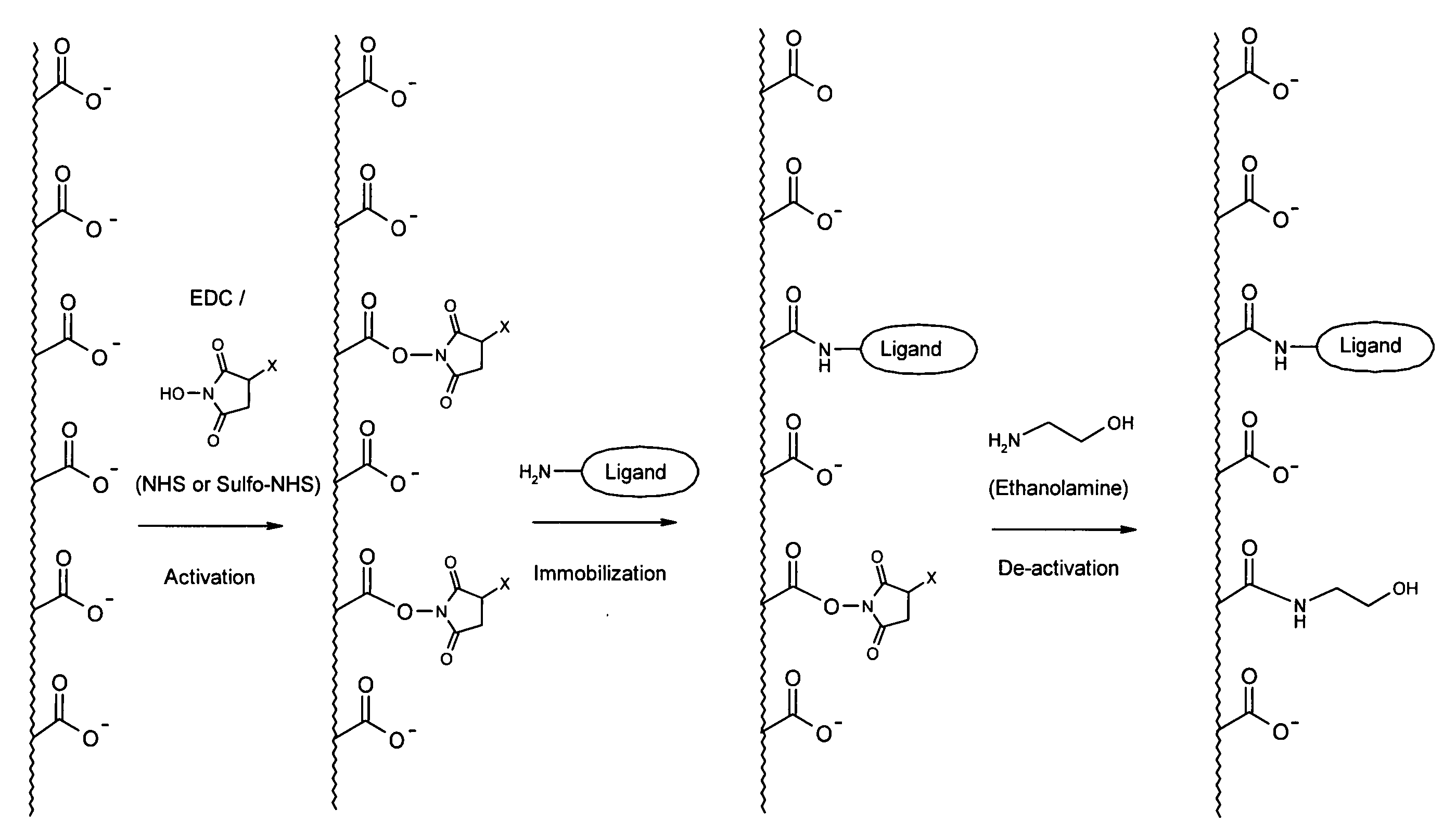
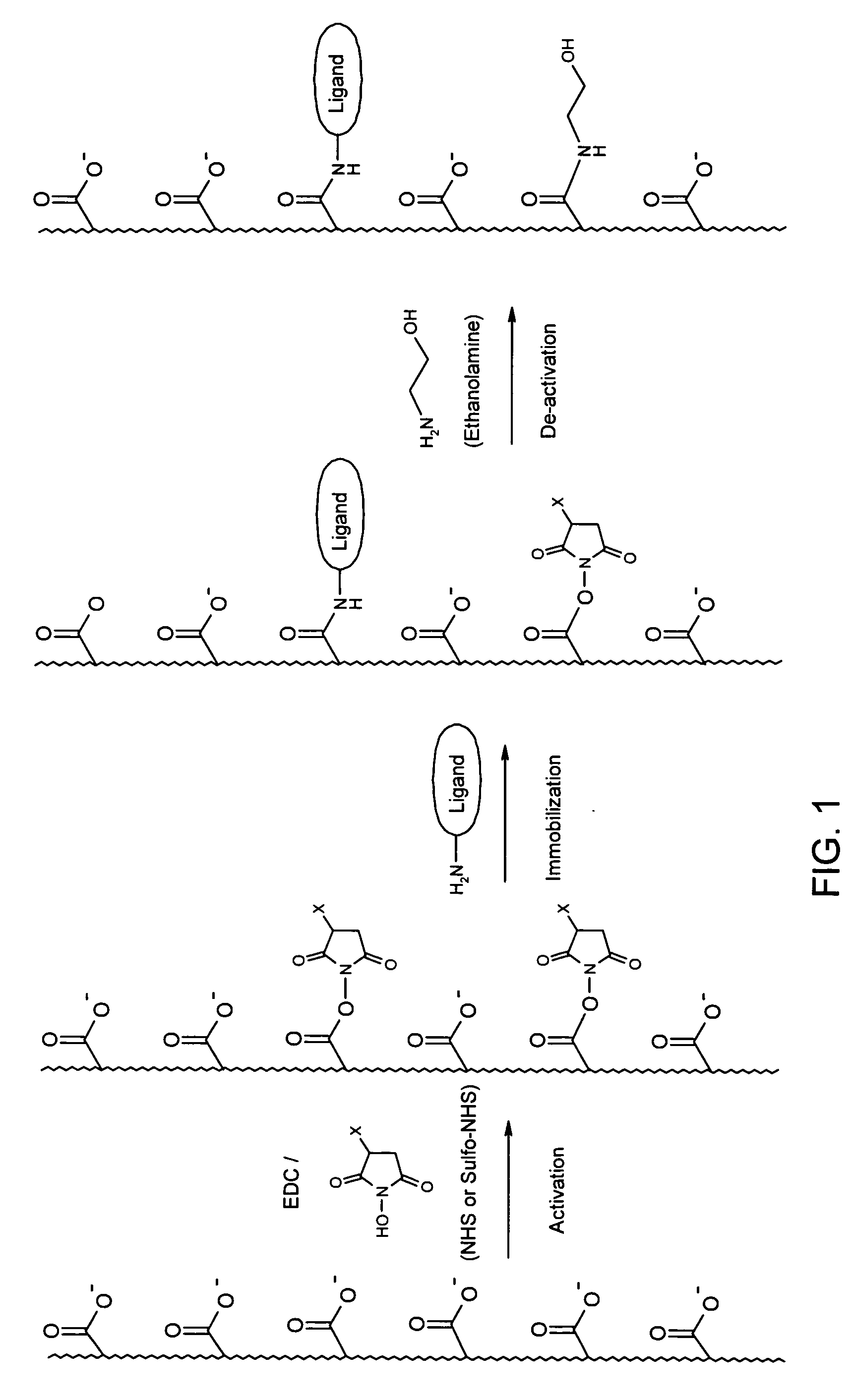
InactiveUS20050176122A1Other chemical processesSolid sorbent liquid separationSpecial classCarboxylic acid
Owner:UPFRONT CHROMATOGRAPHY
Metal-enhanced fluorescence for the label-free detection of interacting biomolecules
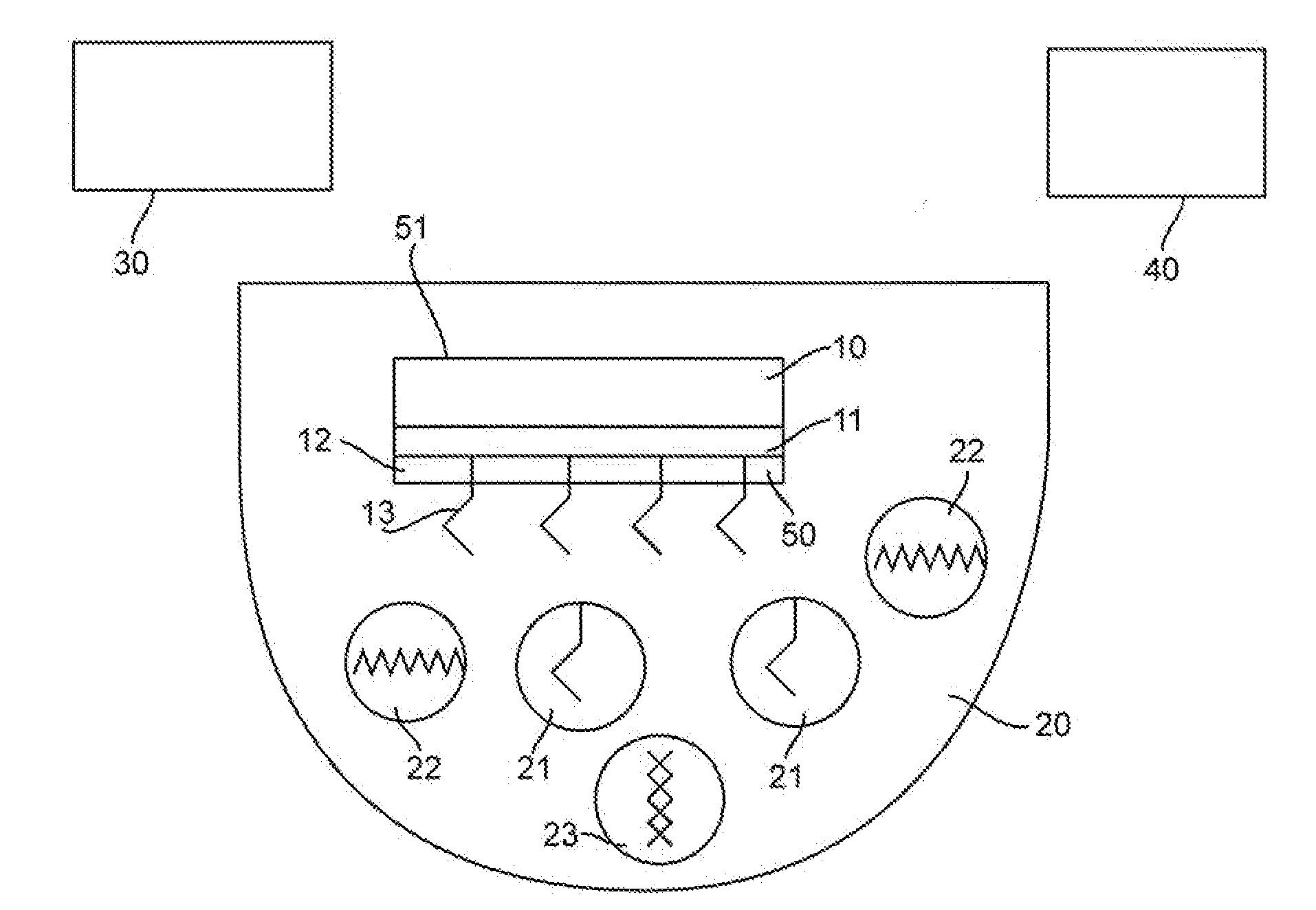
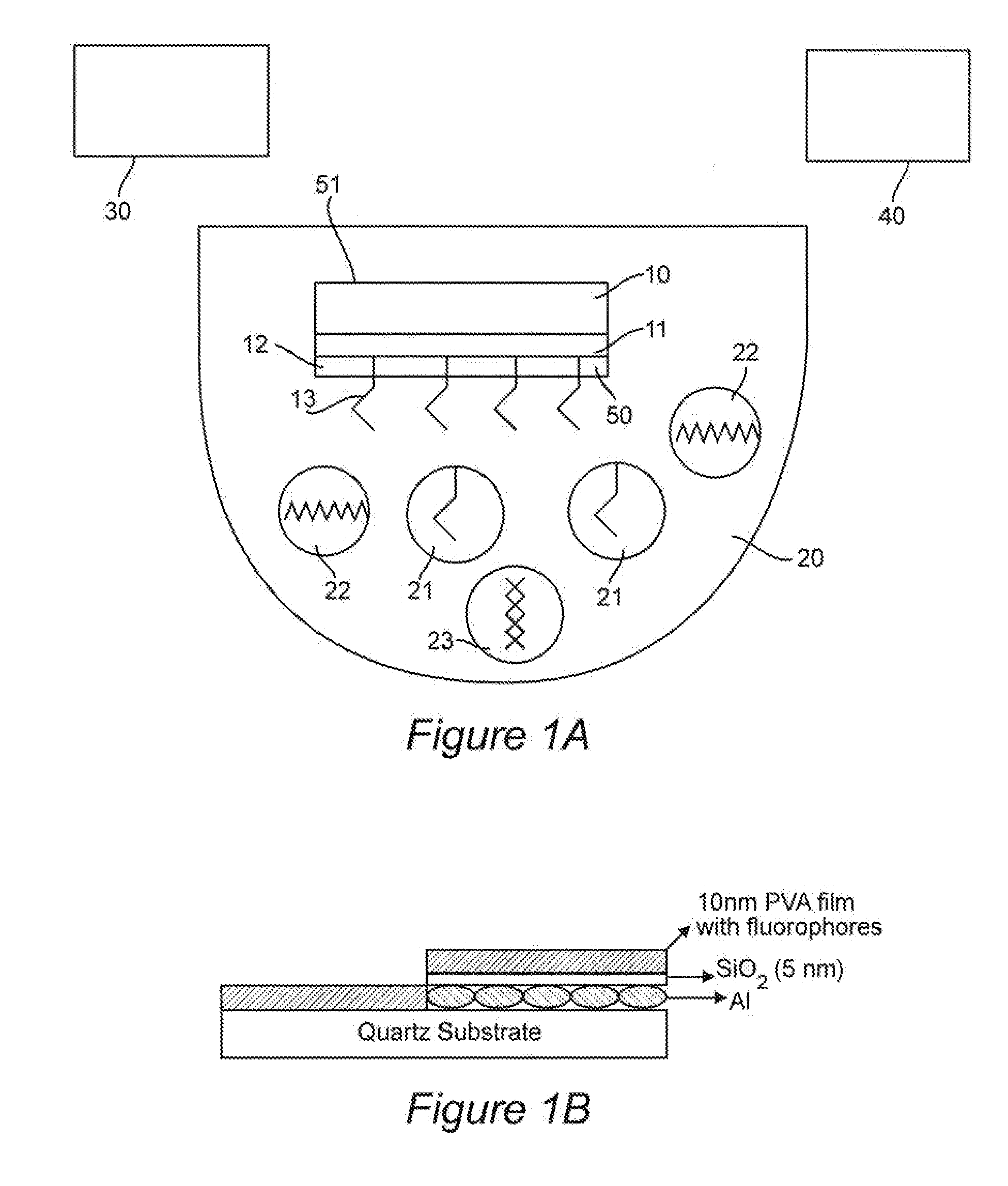
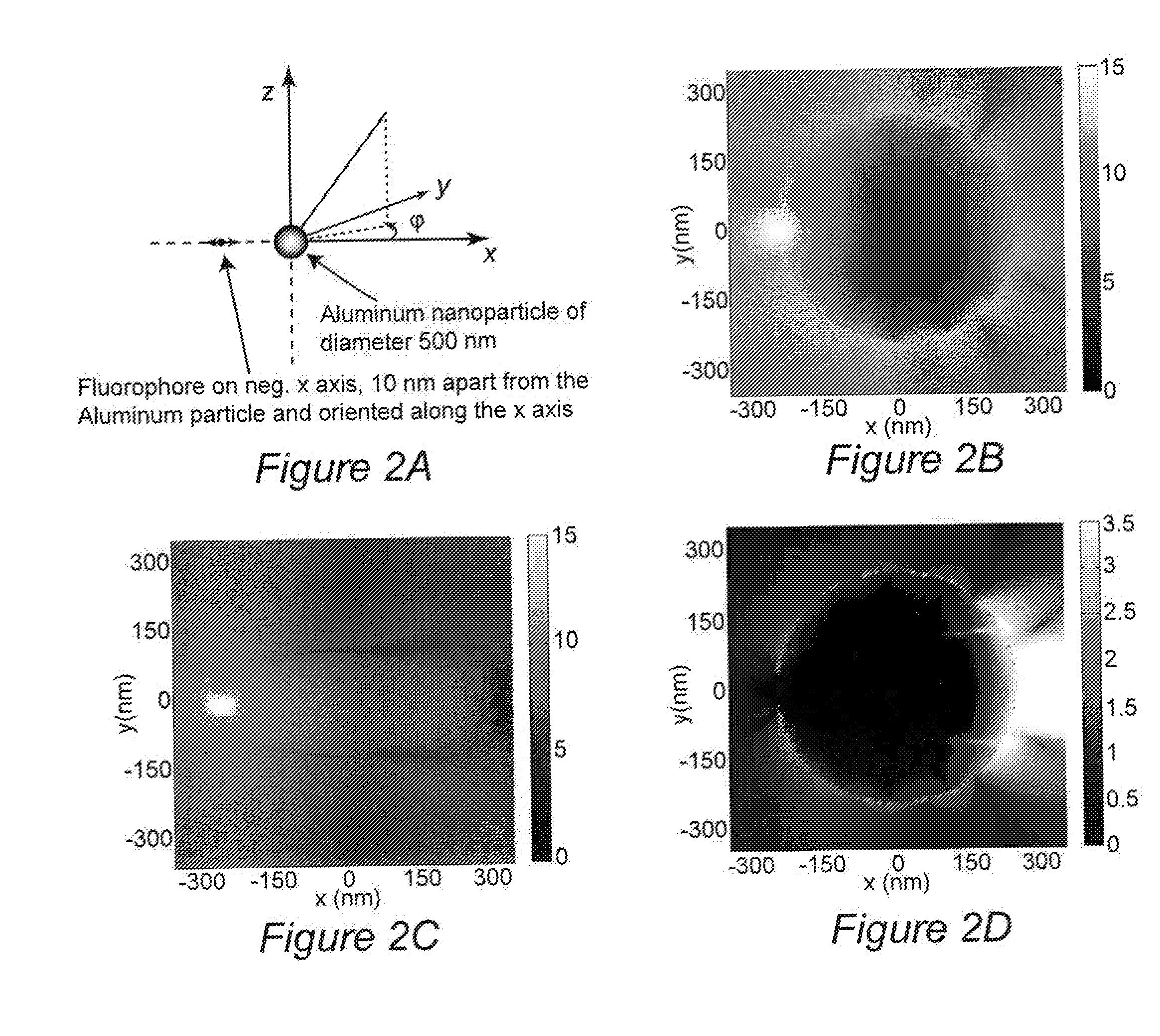
InactiveUS20100035335A1Enhance intrinsic fluorescenceReducing cost and complexityBioreactor/fermenter combinationsBiological substance pretreatmentsNanostructured metalElectromagnetic radiation
A method for enhancing fluorescence of a biomolecule includes the step of associating the biomolecule having intrinsic fluorescence with a sensing surface that contains nanostructured metal. Association of the biomolecule with the nanostructured metal enhances its intrinsic fluorescence, which is detected upon exposure to electromagnetic radiation of a suitable wavelength. The sensing surface may include capture or ligand molecule which binds to the biomolecule and sequesters it in proximity to the nanostructured metal, thereby causing its fluorescent signal to be enhanced. The method can be used in label-free bioassays for detection of interacting biomolecules, such as antibody-antigen binding.
Owner:LAKOWICZ JOSEPH R +4
Targeting viruses using a modified sindbis glycoprotein
InactiveUS20060127981A1High expressionIncrease infectivitySsRNA viruses negative-senseSsRNA viruses positive-senseInfected cellDisease
The present invention relates to viruses that are engineered to contain a surface ligand molecule which targets the virus to a cell of interest. In particular non-limiting embodiments, the cell of interest is desirably ablated and may be a cancer cell, an infected cell, a cell exhibiting a non-malignant proliferative disorder, or a cell of the immune system. Alternatively, the cell of interest is a target for gene therapy.
Owner:UNIVERSITY OF PITTSBURGH
Targeting viruses using a modified sindbis glycoprotein
InactiveUS7429481B2High expressionIncrease infectivitySsRNA viruses negative-senseSsRNA viruses positive-senseNon malignantInfected cell
Owner:UNIVERSITY OF PITTSBURGH
Manufacturing method and readout system for biopolymer arrays
A method for manufacturing an array of biomolecules that have complementary binding species is disclosed. The method utilize a master substrate containing a desired master array of binding molecules or ligands, e.g., DNA target molecules. A mixture of complementary, antiligand molecules, e.g., DNA probe molecules, are then hybridized to the ligands of the master array, Forming an array of antiligand molecules reversibly bound to the molecules in the master array. The antiligand array is then transferred and bound to a separate substrate, to form a desired array of antiligand molecules on the separate substrate. The process may be repeated to produce multiple arrays or multiple generations of arrays. Also disclosed is a sensor array device and method. The device includes an array of vibratory tympanic elements, each having a specific analyte binding probe carried on the vibratory membrane of the element. A binding event is detected by a change in the vibrational resonance of a selected element.
Owner:NEW LIGHT IND
Process for producing beta-diketone functionalization rare earth mesoporous hybridisation luminescent material
InactiveCN101220266ALong fluorescence lifetimeHigh Luminous Quantum EfficiencyLuminescent compositionsDiketoneRare earth
The invention belongs to the technical field of manufacturing nano mesoporous material, in particular to a novel synthesis technology of Beta to diketone functionalized rare earth mesoporous hybrization and luminescent material. The invention first adopts a method of organic modification and synthesis to covalently graft an organic ligand molecule with Beta to diketone structure in a main body framework of an inorganic mesoporous, then rare earth europium or terbium ion is introduced to synthesize the high ordered mesoporous hybrization and luminescent material with steady chemistry and thermodynamic performance. The method and experimenting condition of the invention are moderate, the whole manufacturing system is easy to be established, the operation is easy, and the condition is easy for controlling, so the obtained products has steady quality. .
Owner:TONGJI UNIV
Contrast agent for photoacoustic imaging and photoacoustic imaging method using the same
InactiveUS20110117023A1Sensitive highHigh antigen binding performancePeptide/protein ingredientsSolution deliveryPhotoacoustic imaging in biomedicineVolumetric Mass Density
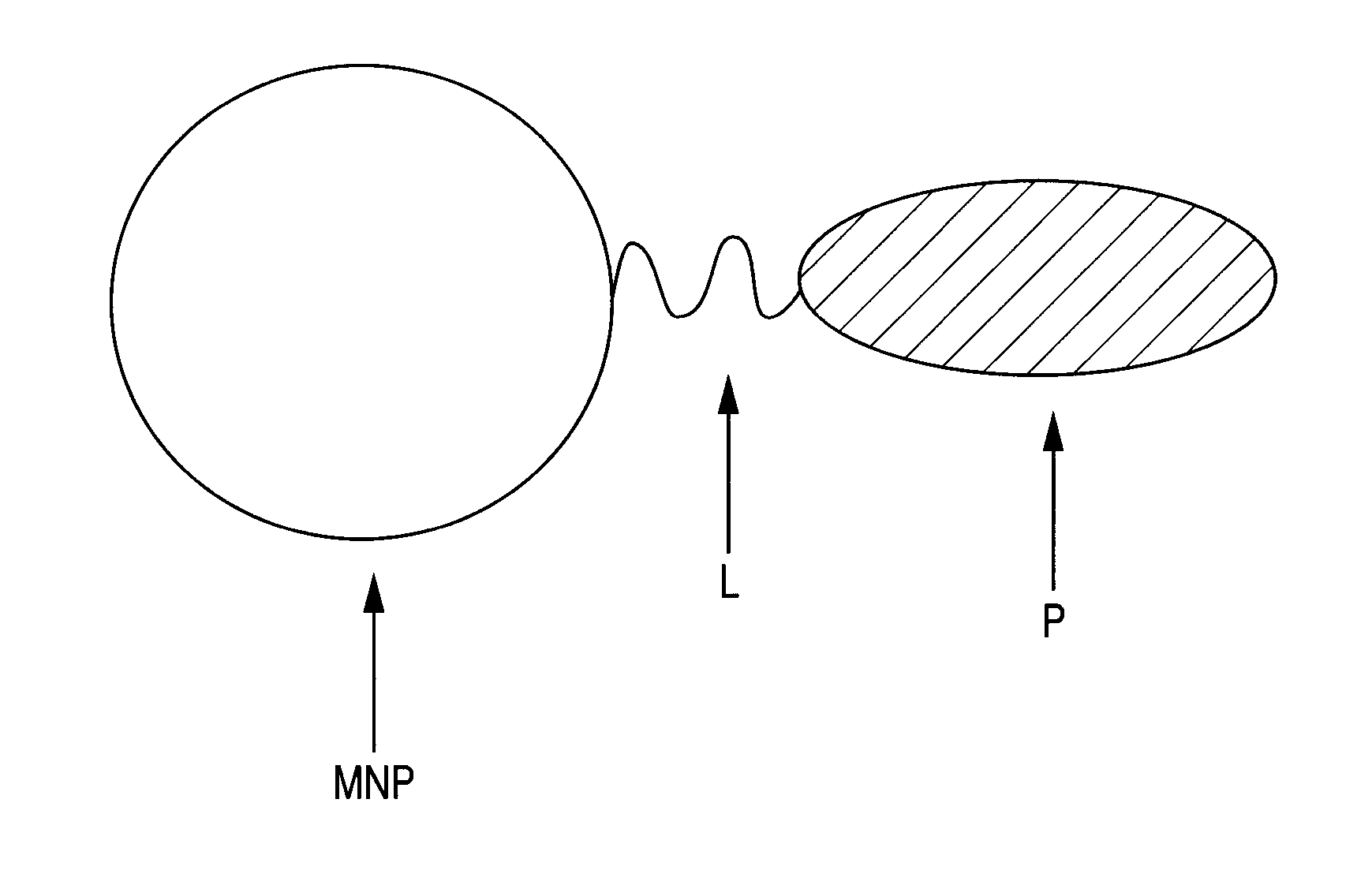
It is intended to provide a novel contrast agent for photoacoustic imaging that is highly capable of binding to a target molecule and generates high photoacoustic signals. The present invention provides a contrast agent for photoacoustic imaging represented by Formula 1:MNP-((L)l-(P)m)n (Formula 1)(wherein MNP represents a particle containing an iron oxide particle; L represents a linker molecule; P represents a ligand molecule; l represents 0 or 1; and m and n represent an integer of 1 or larger), the contrast agent for photoacoustic imaging including: a particle containing an iron oxide particle that absorbs light in a near-infrared region; and at least one or more ligand molecule(s) immobilized on the particle containing an iron oxide particle, wherein the immobilization density of the ligand molecule is equal to or higher than the cell surface density of a target molecule.
Owner:CANON KK
Preparation method of heavy metal ion enrichment cellulosic material
InactiveCN101787654AStrong toughnessHigh mechanical strengthFibrous raw materialsHigh absorptionSilanes
The invention relates to a preparation method of heavy metal ion enrichment cellulosic material; in the method, natural cellulosic material fiber is adopted as the matrix, tetramethyl silane is take as the precursor and the surface sol-gel method is utilized to deposit silica film on the surface of the filter paper fiber; subsequently, ligand molecule monolayer sensitive to copper ion and mercury ion are introduced through self-assembly to obtain the heavy metal ion enrichment cellulosic material; the heavy metal ion solution can be simply filtered by the material prepared by the method of the invention to realize the absorption effect. The heavy metal ion enrichment cellulosic material has superior heavy metal ion enrichment performance, broad raw material source range, low price, simple preparation method, high absorption speed, high sensitiveness and can realize the treatment of waste water containing heavy ions simply, rapidly and economically.
Owner:ZHEJIANG UNIV
Symmetrical difunctional coupling agent and coupled molecular imaging agents thereof
InactiveCN102391168ASolve coupling problemsSolving Difficult-to-Separate PuzzlesIn-vivo radioactive preparationsHybrid peptidesPharmacophoreGastrin-releasing peptide receptor
The invention provides a symmetrical difunctional coupling agent compound (I), i.e. N-fluorenylmethyloxycarbonyl-L-beta-glutamate-di-N-succinimide (Fmoc-beta-Glu(OSu)-OSu), and a production method thereof. Based on the compound, a series of novel coupling compounds which respectively contain a symmetrical difunctional coupling base, i.e. beta-Glu (beta-glutamate) are prepared through being coupled to ligand molecules; the structural formula of the series of coupling compounds is shown as (II), wherein M1 is a -NH2-contained ligand of a target molecule T1, M2 is a -NH2-contained ligand of a target molecule T2, L is a linking group, and S is a report signal group. The compound (1) provided by the invention has been used in coupling a targeted integrin alphavbeta3 receptor ligands, gastrin-releasing peptide receptor ligands, telomerase inhibitor pharmacophores and epidermal growth factor receptor ligands, and thus, a plurality of coupled bi-ligand molecular imaging agents and coupled tri-ligand molecular imaging agents are synthesized. The invention further relates to the use of the compound (I) and the series of novel coupling compounds (II) in the preparation of imaging agent drugs.
Owner:THE FIRST AFFILIATED HOSPITAL OF SUN YAT SEN UNIV
Two-step molecular surface imprinting method for making a sensor
InactiveUS20080179191A1Material analysis by electric/magnetic meansPretreated surfacesThiolMolecular recognition
Owner:MOTOROLA INC
Furin-cleavable peptide linkers for drug-ligand conjugates
InactiveUS20090155289A1Targeted optimizationGood target for cancer cellsPeptide/protein ingredientsImmunoglobulins against animals/humansTrans golgiLigand molecule
Disclosed are certain peptide linkers for conjugating drugs to ligands, and the resulting drug-linker-ligand molecules and compositions thereof. The conjugated molecules useful for the targeted delivery of drugs to the desired cells, and allow for the intracellular release of the drug in cases where the targeted antigen is internalized via the trans Golgi network and not the lysosomal pathway.
Owner:PANACEA PHARMA
Catalytic system for obtaining conjugated diene/monoolefin copolymers and these copolymers
InactiveUS20060160969A1Organic-compounds/hydrides/coordination-complexes catalystsCatalyst activation/preparationGrignard reagentLanthanide
A catalytic system usable for the copolymerization of at least one conjugated diene and at least one monoolefin, a process for preparing this catalytic system, a process for preparing a copolymer of a conjugated diene and at least one monoolefin using said catalytic system, and said copolymer are described. This catalytic system includes: (i) an organometallic complex represented by the following formula: {[P(Cp)(Fl)Ln(X)(Lx)}p (1) where Ln represents a lanthanide atom to which is attached a ligand molecule comprising cyclopentadienyl Cp and fluorenyl Fl groups linked to one another by a bridge P of the formula MR1R2, M is an element from column IVa of Mendeleev's periodic table and R1 and R2 each represent alkyl groups of 1 to 20 carbon atoms or cycloalkyl or phenyl groups of 6 to 20 carbon atoms, X represents a halogen atom, L represents an optional complexing molecule, such as an ether, and optionally a substantially less complexing molecule, such as toluene, p is a natural integer greater than or equal to 1 and x is greater than or equal to 0, and (ii) a co-catalyst selected from alkylmagnesiums, alkyllithiums, alkylaluminums, Grignard reagents and mixtures of these constituents.
Owner:MICHELIN RECH & TECH SA +1
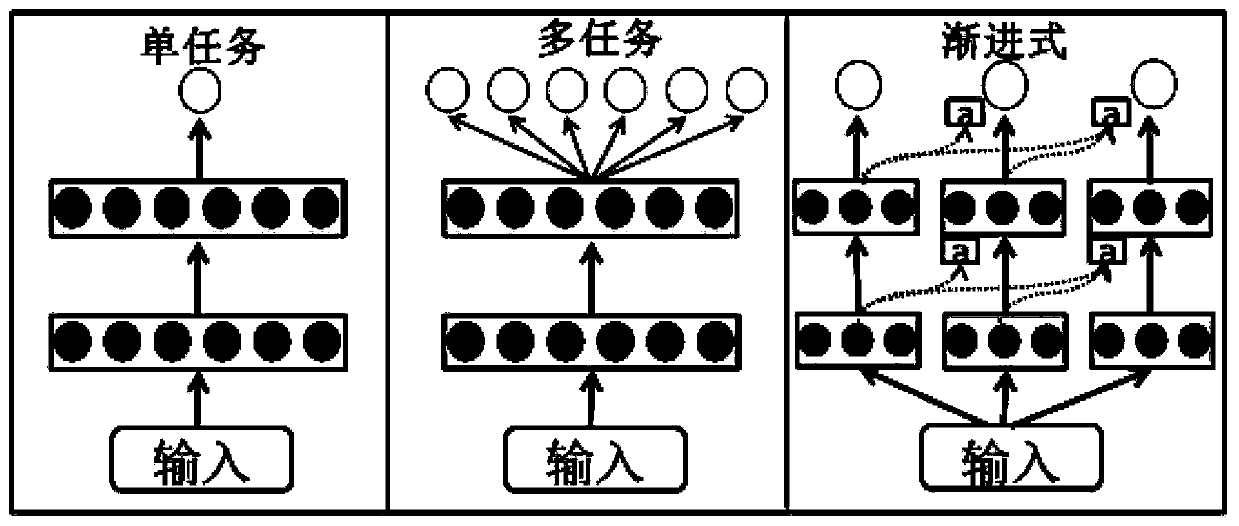
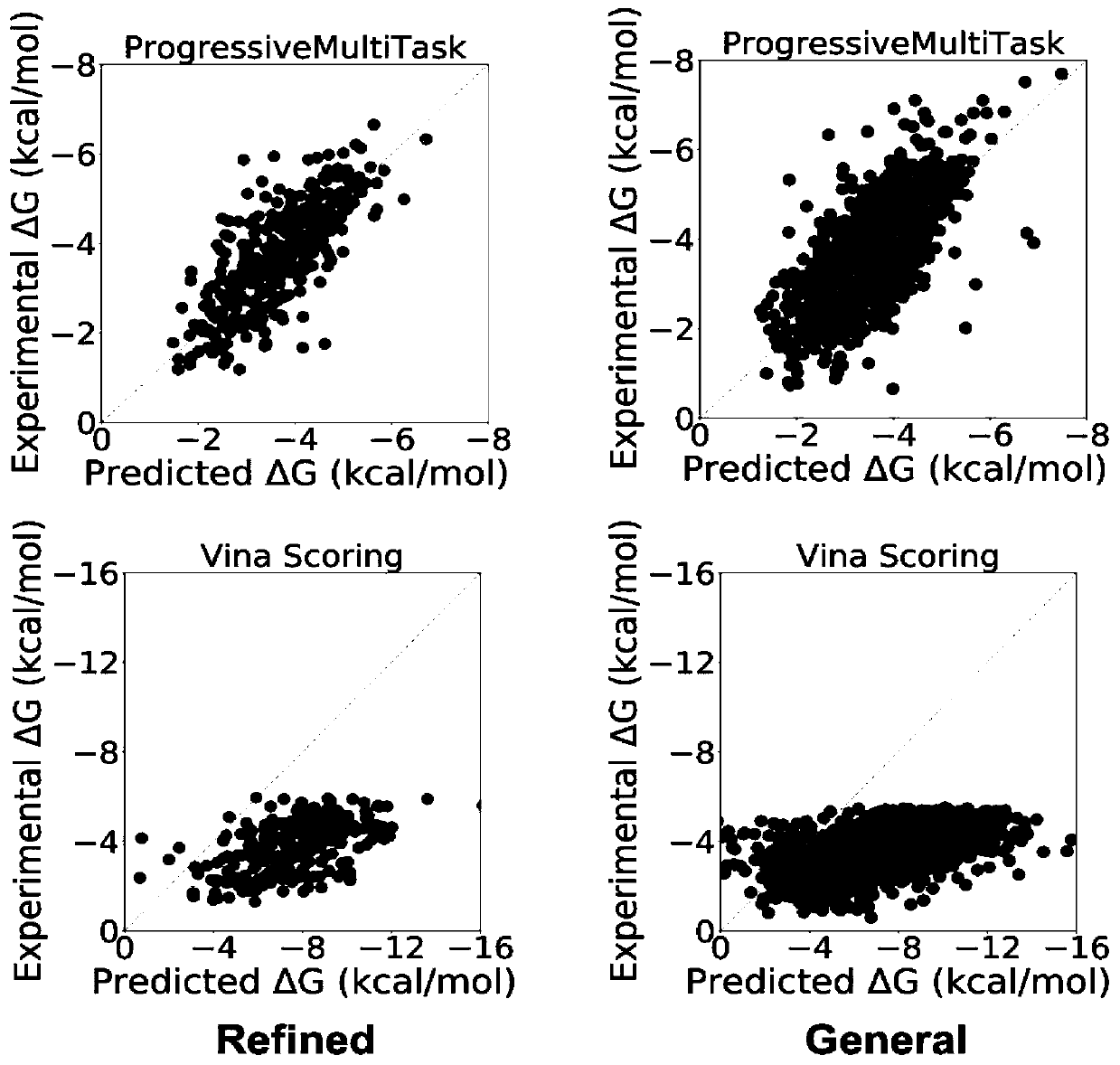
Method for predicting binding free energy of protein and ligand based on progressive neural network
ActiveCN110910951APrevent overfittingImprove generalization abilityProteomicsGenomicsAlgorithmTest set
The invention discloses a method for predicting the binding free energy of protein and ligand based on a progressive neural network, and belongs to the technical field of computer-aided drug design. The method comprises the steps: obtaining a pdb file from a PDBbind database, establishing local database, acquiring an amino acid molecule within 4.5 angstroms in the protein binding pocket by takingthe ligand molecule as a center, performing extended connectivity fingerprint calculation, carrying out SPLIF fingerprint calculation, searching for the number of salt bridges and hydrogen bonds between protein and ligand molecules, converting the structural information of the protein and the ligand into a one-dimensional tensor, and establishing a training set, a verification set and a test set;training the progressive neural network by using the training set; optimizing and searching hyper-parameters for prediction; through comparison with a molecular docking result, obtaining a higher Pearson correlation coefficient. According to the invention, the technical problem of how to convert a three-dimensional structure of protein and ligand molecules into tensors which are easy to calculateby a computer and input the tensors into the progressive neural network for training and optimization is solved, and the calculation rate and the prediction accuracy are greatly improved.
Owner:JIANGSU UNIV OF TECH
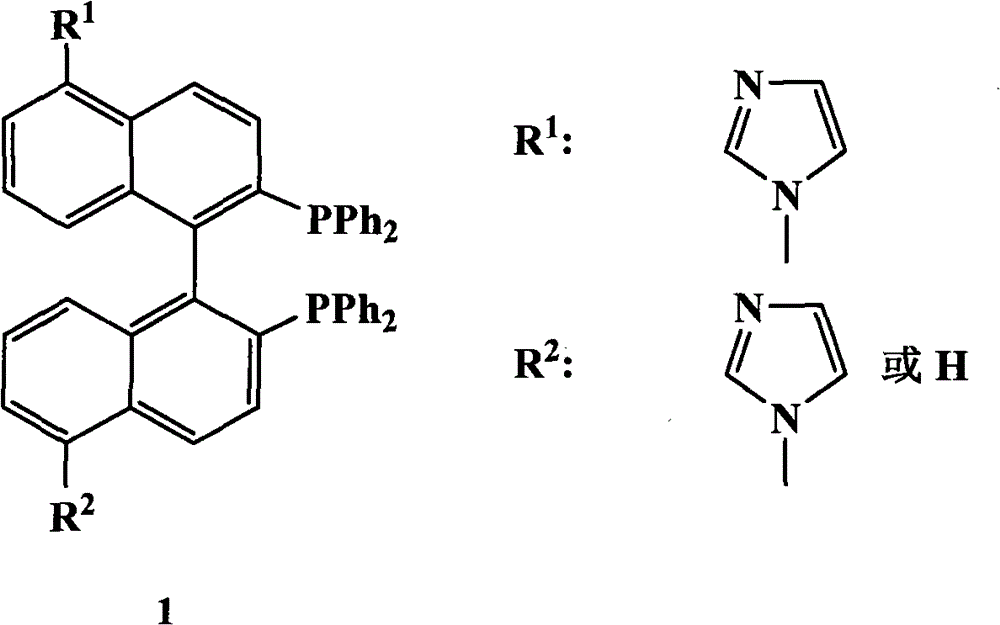
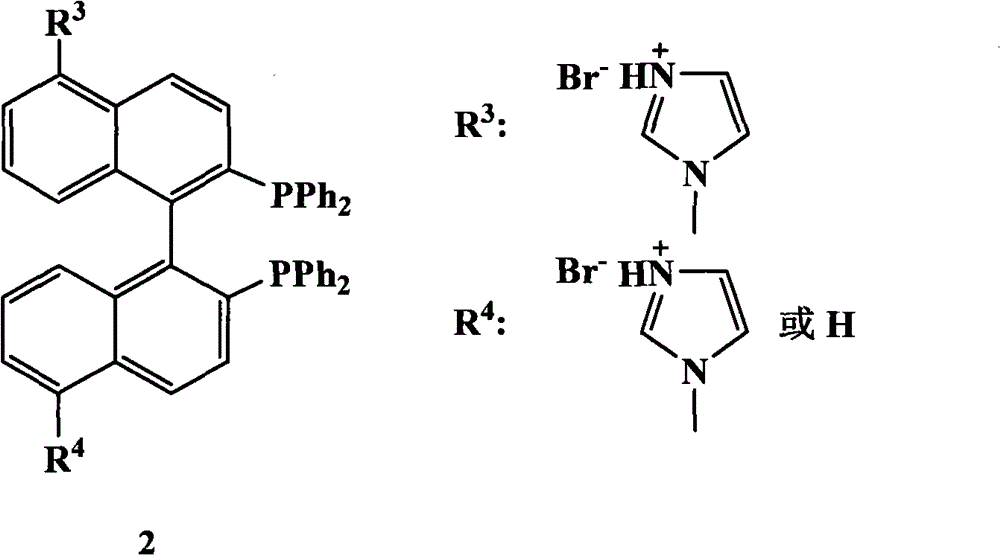
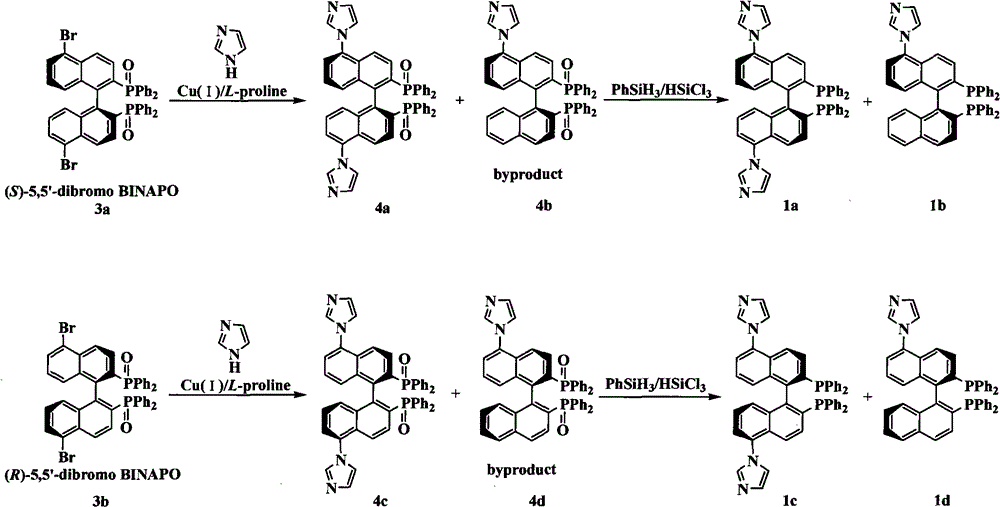
Chiral diphosphine ligand and chiral catalyst, and preparation and application method thereof
ActiveCN103059064AHigh affinityReduced activityOrganic compound preparationGroup 5/15 element organic compoundsCyclic processDiphosphines
The invention relates to a chiral diphosphine ligand and a chiral catalyst, and a preparation and application method thereof. The preparation method of a BINAP (2,2'-bis(diphenylphosphino)-1,1'-binaphthyl) chiral diphosphine ligand based on novel imidazole and imidazole cation modification. Imidazole and imidazole cation are introduced to the 5,5'- position of the BINAP molecule framework to synthesize the chiral diphosphine ligand. The assembly of the imidazole and imidazole cation into the chiral diphosphine ligand molecule can effectively enhance the stability of the chiral catalyst in an ionic liquid and the affinity with the imidazole ionic liquid, and avoids loss of the catalyst in the cyclic process. The imidazole-modified chiral diphosphine ligand has the following chemical structural formula, wherein the spatial configuration of the imidazole-modified chiral diphosphine ligand is S type or R type.
Owner:山东聚强绿洲生物科技有限公司
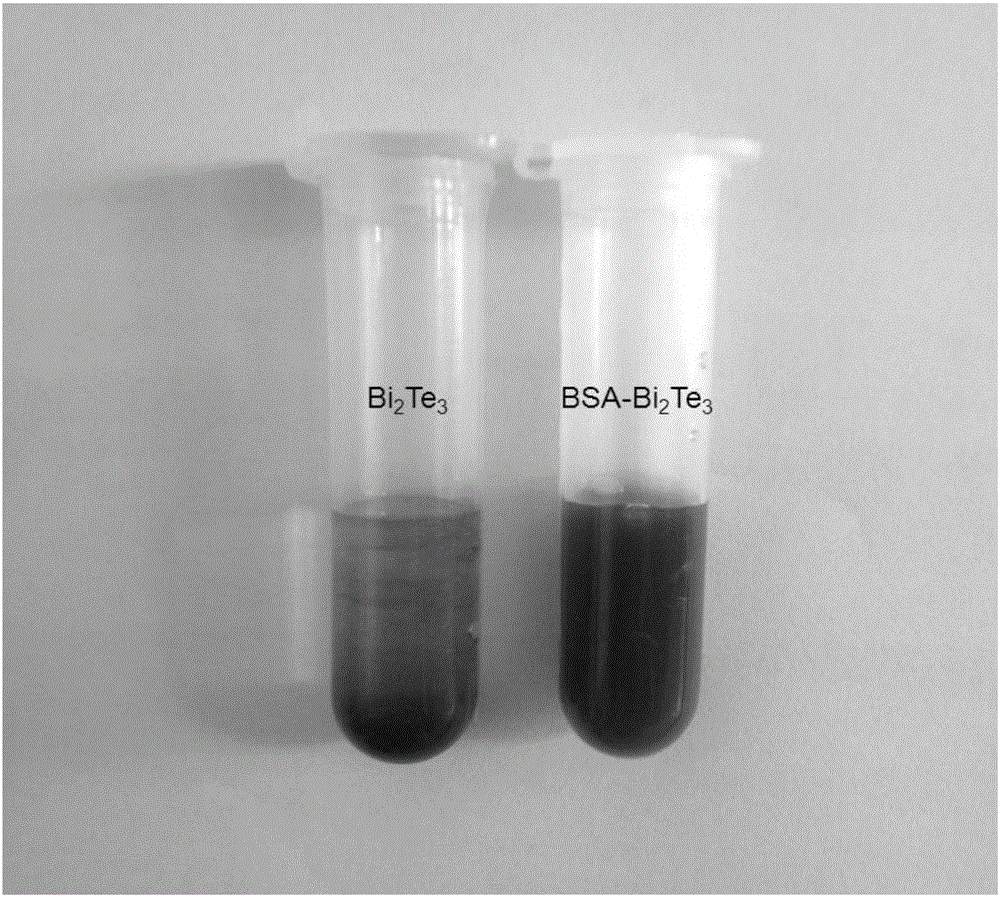
Bi2Te3 dimensional nano tablet, preparation method and applications thereof
ActiveCN105060259AImprove stabilityGood biocompatibilityMaterial nanotechnologyBinary selenium/tellurium compoundsSolubilityBiocompatibility Testing
The invention provides a preparation method of a Bi2Te3 dimensional nano tablet. The preparation method comprises a step of subjecting a Bi2Te3 nano material, water, and functional ligand molecules to ultrasonic exfoliation so as to obtain the Bi2Te3 dimensional nano tablet; wherein the functional ligand molecule is polyvinylpyrrolidone, cytochrome C, or bovine serum albumin. In the preparation method, the functional ligand molecule is taken as the stripping agent to carry out ultrasonic exfoliation on a Bi2Te3 nano material so as to obtain a functional ligand molecule-Bi2Te3 dimensional nano tablet with high water solubility; and the dimensional nano tablet has the advantages of uniform particle size, controllable morphology, high water solubility, good biocompatibility, and high photo-thermal conversion efficiency, is a good photo-thermal agent, and can be applied to tumor photo-thermal therapy. The provided preparation method can synthesize massive water-soluble Bi2Te3 with a photo-thermal property, the preparation process is simple, the cost is low, and the preparation method is suitable for industrial production.
Owner:JIANGXI INST OF RARE EARTHS CHINESE ACAD OF SCI
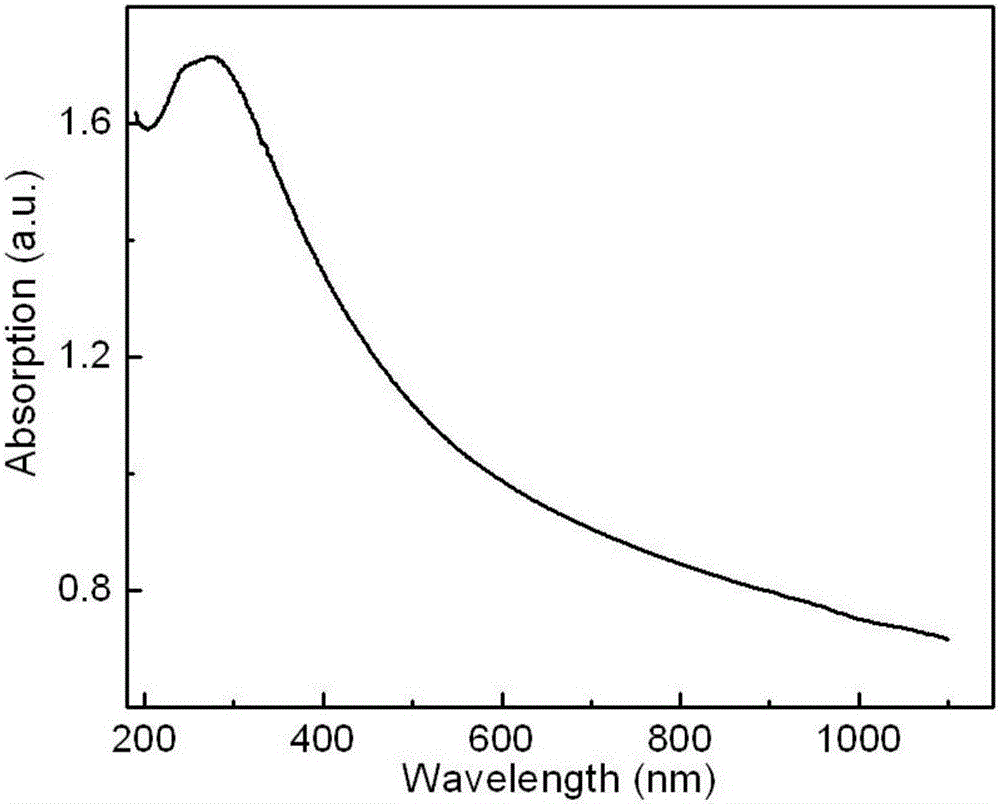
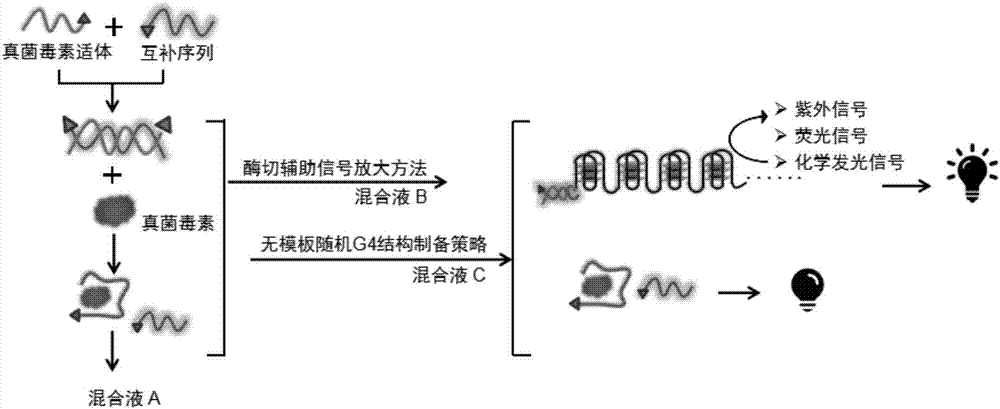
Method for detecting fungaltoxin through multiple signals and kit
ActiveCN107271668ARich identification methodsRich Signal TransformationBiological material analysisBiological testingFluorescenceUltraviolet
The invention discloses a method for detecting fungaltoxin through multiple signals. The method comprises the following steps: 1) combining fungaltoxin with aptamer: adding a to-be-detected sample after the reaction of the fungaltoxin aptamer and complementary sequence thereof, thereby acquiring a mixed solution A; 2) amplifying a digestion auxiliary signal: reacting the mixed solution A with restriction enzyme, thereby acquiring a mixed solution B; 3) preparing a guanine tetramer structure: reacting the mixed solution B with tail-end deoxynucleotide transferase and deoxyribonucleoside triphosphate and reacting with ligand molecules, thereby acquiring a mixed solution C; and 4) detecting and analyzing: performing catalytic oxidation reaction on the mixed solution C and different substrates, generating ultraviolet, fluorescent and chemiluminescent signals and calculating the content of fungaltoxin in the to-be-detected sample according to the relation of the response strength of all the signals and the fungaltoxin concentration. The method disclosed by the invention has the characteristics of quick and simple operation and treatment, short detection time, marker-free effect, low cost, high precision, high sensitivity, and the like.
Owner:ACAD OF NAT FOOD & STRATEGIC RESERVES ADMINISTRATION
Method and apparatus for bio-molecular chip minute quantity sample application and reaction
InactiveCN1523354AUniform fixationUniform reactivitySequential/parallel process reactionsMicrobiological testing/measurementContact formationEngineering
The invention relates to a method to directly add trace sample and instantly react on the biomolecule chip and device. The method includes: under the action of external pressure, the surfaces of microarray and smooth chips close contact to form a sealed cavity. The bio-ligand molecule which needs fixing enters the cavity through the micro-flow channel and reacts with the chip surfaces, and after reacting, the ligand molecule is discharged through the micro-flow channel. The device includes: elastic material slice carved with grooves, microarray arranged of grooves and micro-flow channel matching the microarray; and elastic material membrane covered on the micro-flow channel, where there is a liquid entrance made on the base slice of microarray or micro-flow channel. After fixed and reacting, the chip surfaces are washed by buffer liquor, to make fixation and reaction of the biomolecule on the surface uniform and consistent, effectively heightening the detecting quality.
Owner:INST OF MECHANICS - CHINESE ACAD OF SCI
NMR nuclear magnetic resonance method for high flux medicine screening
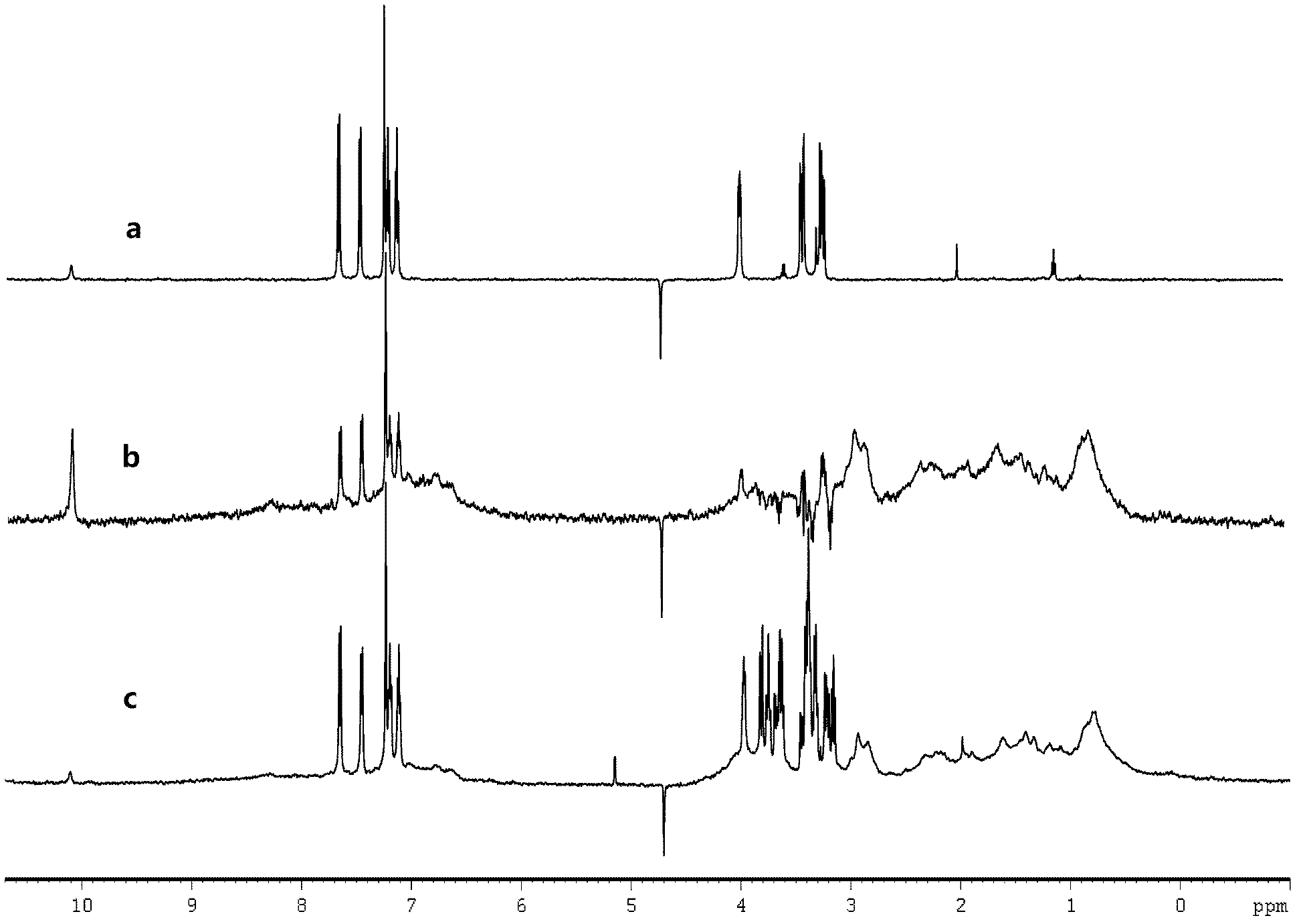
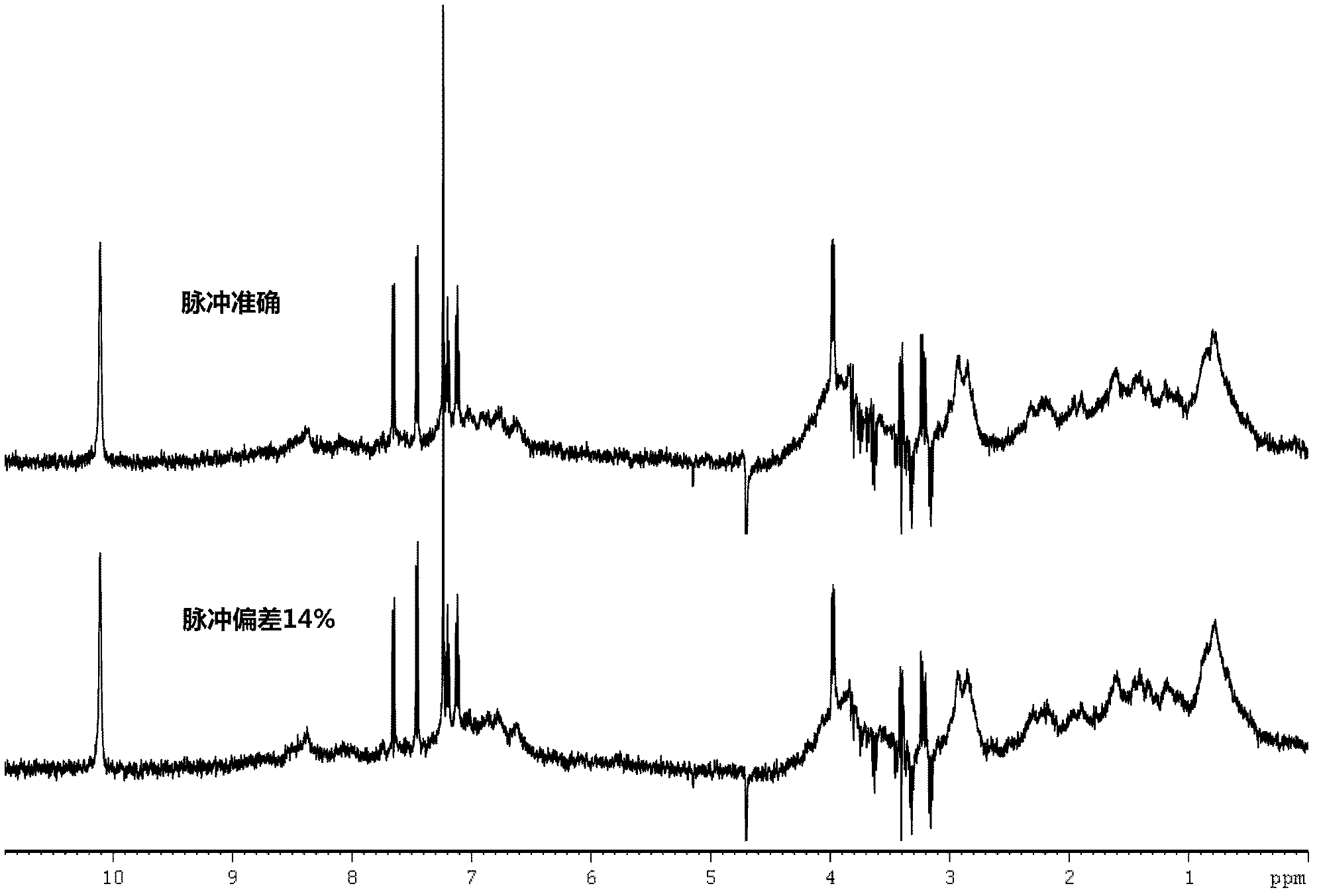
ActiveCN102507627AHigh sensitivityAvoid relaxationWater resource assessmentAnalysis using nuclear magnetic resonanceNMR - Nuclear magnetic resonanceProtein target
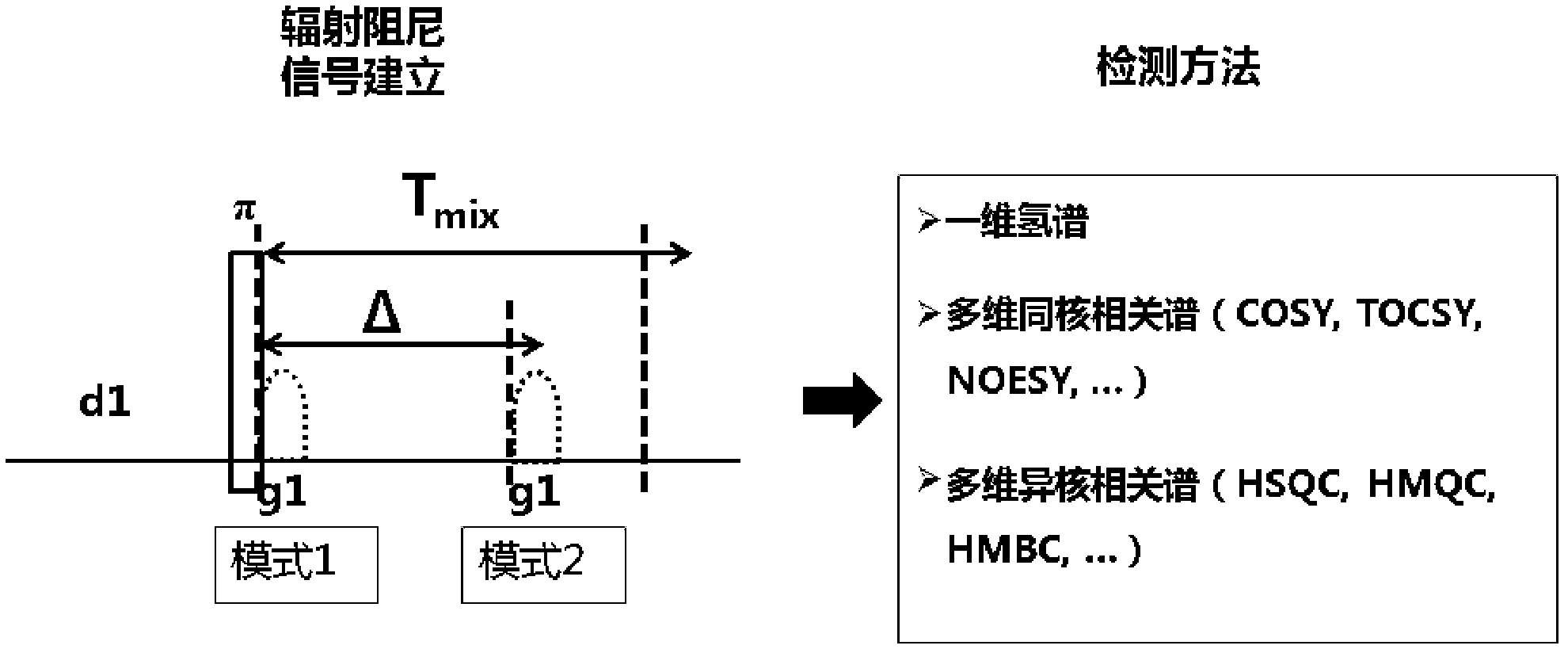
The invention discloses an NMR nuclear magnetic resonance method for high flux medicine screening. The method includes the following steps: a, preparing mixed water solution containing target biomacromolecules and to-be-screened medicine, and adding the target biomacromolecules and to-be-screened molecules in the water solution and uniformly mixing the biomacromolecules and the to-be-screened molecules as well as the water solution; b, filling the mixed solution in an NMR sample tube, placing the sample tube in a spectrometer, and selectively detecting signals of ligand molecules combined with the target protein, wherein the selective detection includes the steps that firstly, building the selective disturbance for water serving as the resolvent based on the radiation damping effect, secondly, performing the operation based on the effects such as exchange and polarization transfer, and thirdly, exciting the built signals, and selectively detecting the protein and the ligand molecules of the protein, wherein the operation includes two pulse modular units, namely, a signal building module and a detection module; and c, completing the molecule confirmation. The method is simple and convenient to operate and feasible, has high accuracy and sensitivity in experiment detection, and effectively improves the accuracy and efficiency in medicine screening.
Owner:INNOVATION ACAD FOR PRECISION MEASUREMENT SCI & TECH CAS
Catalytic system for obtaining conjugated diene/monoolefin copolymers and these copolymers
InactiveUS7547654B2Organic-compounds/hydrides/coordination-complexes catalystsCatalyst activation/preparationPolymer sciencePtru catalyst
A catalytic system usable for the copolymerization of at least one conjugated diene and at least one monoolefin, a process for preparing this catalytic system, a process for preparing a copolymer of a conjugated diene and at least one monoolefin using said catalytic system, and said copolymer are described. This catalytic system includes:(i) an organometallic complex represented by the following formula:{[P(Cp)(Fl)Ln(X)(Lx)}p (1)where Ln represents a lanthanide atom to which is attached a ligand molecule comprising cyclopentadienyl Cp and fluorenyl Fl groups linked to one another by a bridge P of the formula MR1R2, M is an element from column IVa of Mendeleev's periodic table and R1 and R2 each represent alkyl groups of 1 to 20 carbon atoms or cycloalkyl or phenyl groups of 6 to 20 carbon atoms, X represents a halogen atom, L represents an optional complexing molecule, such as an ether, and optionally a substantially less complexing molecule, such as toluene, p is a natural integer greater than or equal to 1 and x is greater than or equal to 0, and(ii) a co-catalyst selected from alkylmagnesiums, alkyllithiums, alkylaluminums, Grignard reagents and mixtures of these constituents.
Owner:MICHELIN RECH & TECH SA +1
Light-sensitive luminous europium complex and light-sensitive ligand molecular and their synthesizing method
InactiveCN101070318AExcellent long-wave sensitization and luminescence abilityWide excitation windowGroup 3/13 element organic compoundsLuminescent compositionsFluorescenceStructural formula
This invention relates to a photosensitive shining Europium complex and its photosensitive ligand molecule, also relates to the synthetic method. The Europium complex structure is showd as VI: the R1, R2 is alkyl with 1 to 4 carbon atoms, R3, R4, R5, and R6 are same and are methyl or H, the R8 is methyl or H. The organic photosensitive ligand molecule structural formula is showd as I: the R1, R2 is alkyl with 1 to 4 carbon atoms, R3, R4, R5, and R6 are same and are methyl or H, the R8 is methyl or H. The photoallergy molecule possess fine long wave photoactivate Europium ion luminance. The new style Europium complex possess width activating window, at the same time possess large two-photon absorption acting section and / or high rare earth ion fluorescence quanta yield, is useful for preparing excellent bioluminescence probe stuff, possess comprehensive application prospects.
Owner:北京纳材科技有限公司
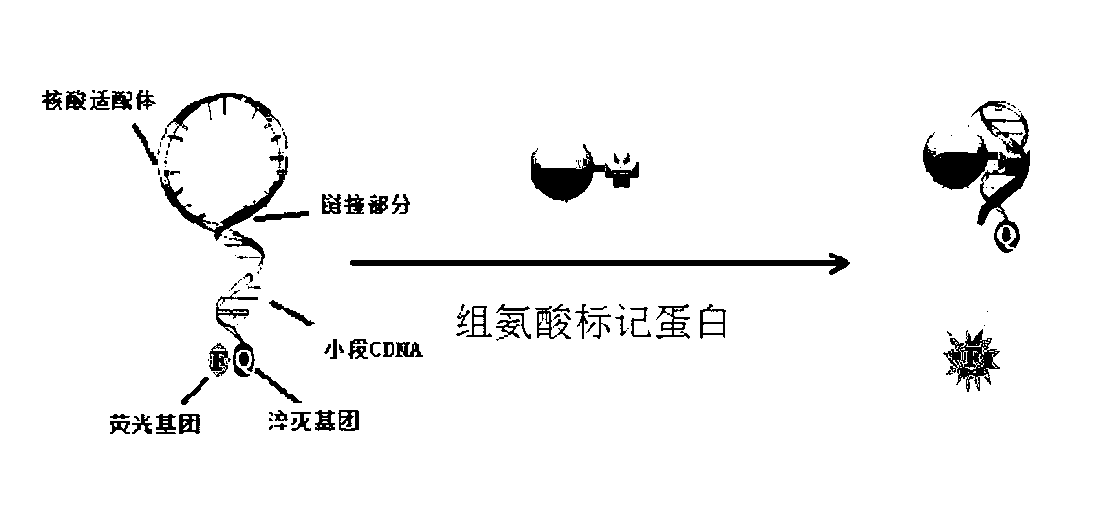
Nucleic acid aptamer molecular beacon probe for detecting histidine-tag recombinant proteins and detection method thereof
ActiveCN102719430ARapid detection and quantitative analysisHigh affinityMicrobiological testing/measurementFluorescence/phosphorescenceProtein markersPolyhistidine-tag
The invention discloses a nucleic acid aptamer molecular beacon probe for detecting histidine-tag recombinant proteins and a direct general method for detecting the histidine-tag recombinant proteins in cell lysates by the probe. The nucleic acid aptamer probe comprises sequences shown in SEQIDNO.1 and SEQIDNO.2; the end 3' of the sequence shown in the SEQIDNO.1 is bridged with the end 5' of the sequence shown in the SEQIDNO.2 through PEG36; the end 5' of the sequence shown in the SEQIDNO.1 is modified with fluorescein FAM; and the end 3' of the sequence shown in the SEQIDNO.2 is modified with a quenching group Dabcy1. According to the nucleic acid aptamer molecular beacon probe, the expression of the histidine-tag recombinant proteins in the cell lysates can be quickly and quantificationally analyzed without protein tagging, and the method disclosed by the invention has the advantages of universality and low cost, and the advantage that the histidine-tag recombinant proteins can be simply, quickly and quantificationally analyzed.
Owner:HUNAN UNIV
Method for preparing silver sulfide quantum dot using MOFs as carrier
InactiveCN107880876AGood light stabilityGood chemical stabilityMaterial nanotechnologyNanoopticsChemical stabilityComputational chemistry
The invention discloses a method for preparing a silver sulfide quantum dot using MOFs as a carrier. The method uses the MOFs as the carrier and a template, silver nitrate or silver acetate as a silver source, sodium sulfide as a sulfur source, and a hydrazone-containing hydrophilic reagent as a surface ligand molecule, and achieves one-step preparation of the silver sulfide quantum dot with the MOFs as the carrier in an aqueous phase system. The prepared silver sulfide quantum dot with the MOFs as the carrier has an emission wavelength between 900 and 1250 nm, has good near-infrared fluorescence and chemical stability, and can be applied to the fields of bioluminescent labeling, imaging and photoelectric devices.
Owner:苏州影睿光学科技有限公司
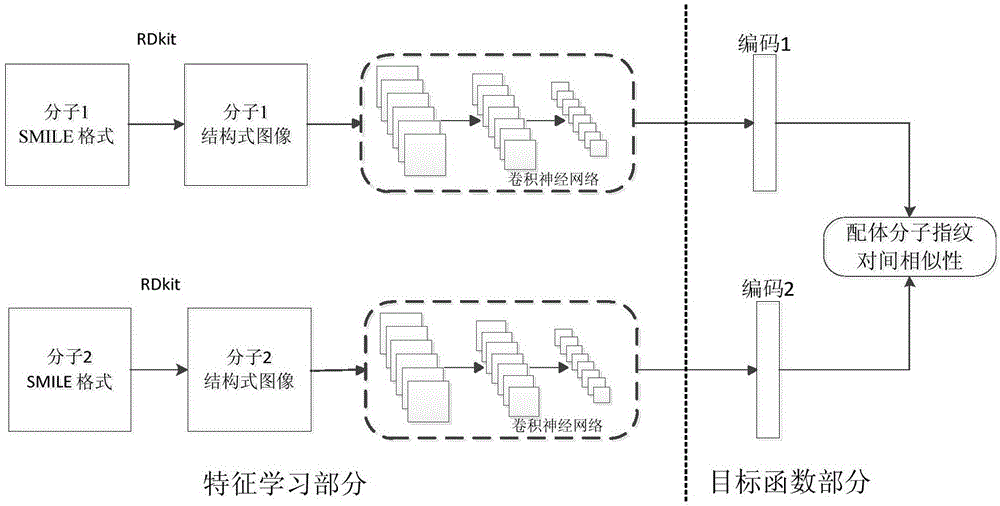
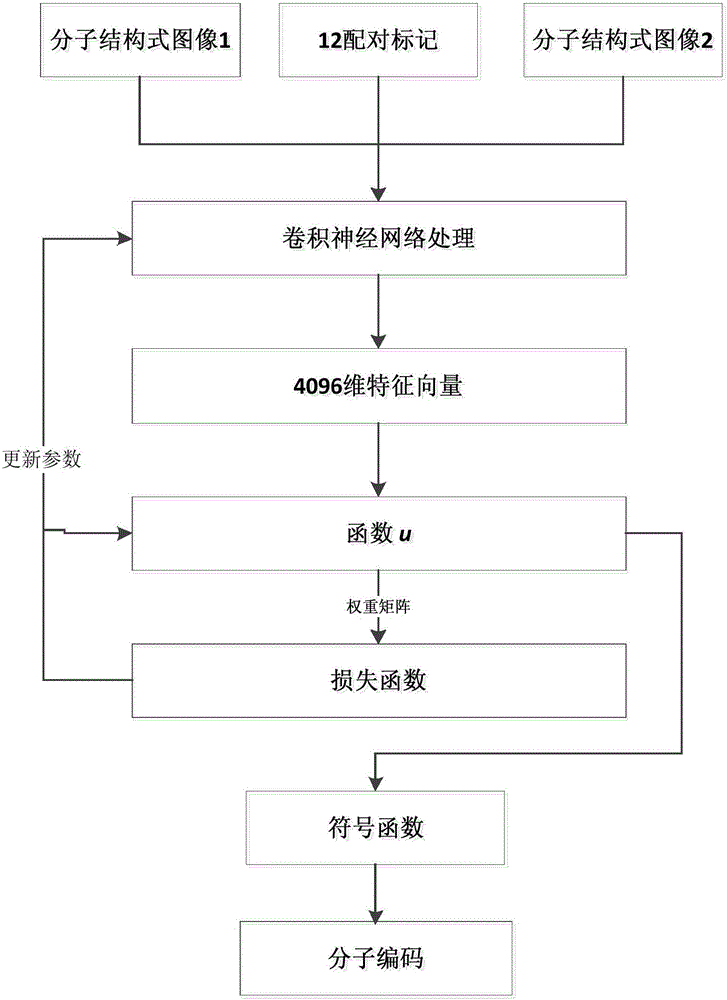
Ligand molecular fingerprint generation method based on deep Hash in drug screening
ActiveCN106777986AChemical property predictionMolecular entity identificationAlgorithmStructural formula
The invention discloses a ligand molecular fingerprint generation method based on deep Hash in drug screening. Firstly, a molecular structural formula image file is generated. Secondly, a pairwise label of a ligand molecular pair is defined, and a DPSH deep Hash learning model is trained. Finally, molecular fingerprint of new ligand molecules is predicted. A ligand molecular structural formula is converted into the image file, a target loss function is optimized by a deep Hash algorithm, and the molecular fingerprint is automatically generated. A first 'end-to-end' molecular fingerprint generation framework can be realized without manually extracting characteristics, and the method solves the problem that developers need to deeply understand field knowledge in an existing molecular fingerprint generation method. The universal molecular fingerprint generation framework is provided from a new perspective and serves as an important supplement for the existing molecular fingerprint generation method, and wider application of the molecular fingerprint to drug screening can be promoted.
Owner:NANJING UNIV OF POSTS & TELECOMM
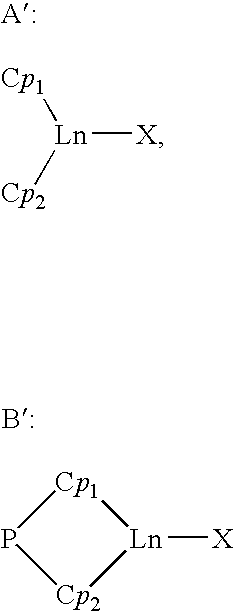
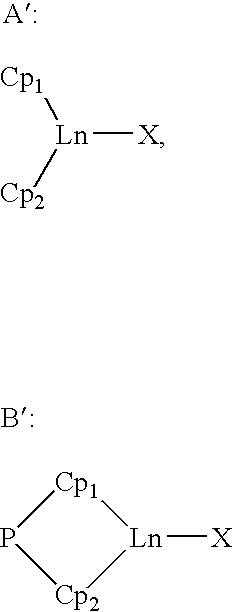
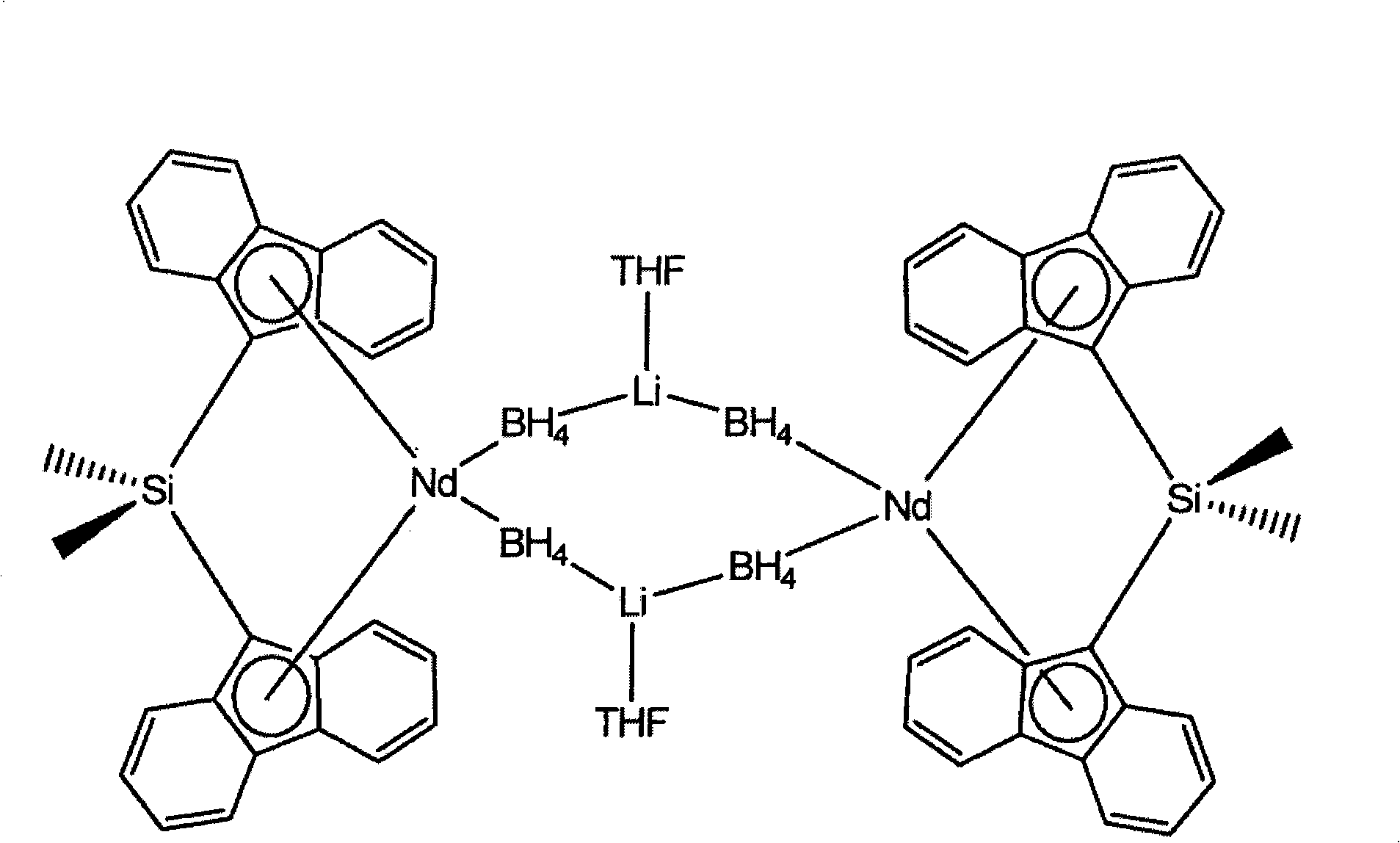
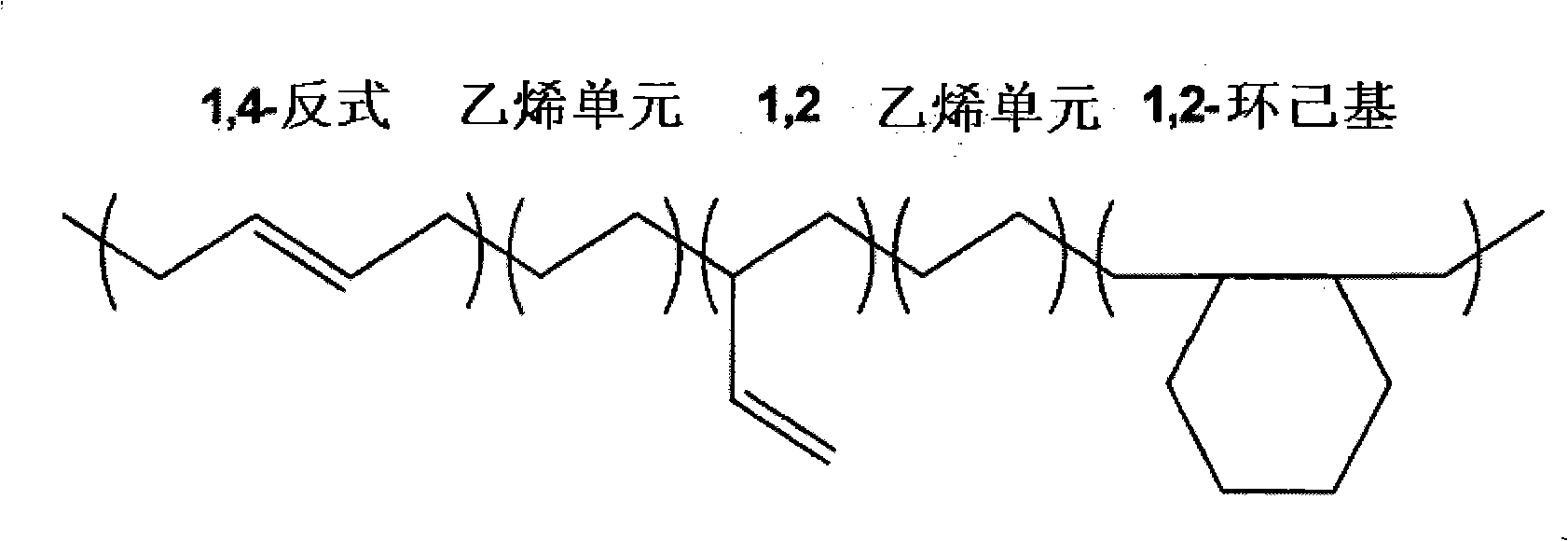
Borohydride metallocene complex of a lanthanide, catalytic system including said complex, polymerization method using same and ethylene/butadiene copolymer obtained using said method
The invention provides a borohydride metallocene complex of a lanthanide, a preparation method thereof, a catalytic system including said borohydride metallocene complex, a method for the copolymerisation of olefins using said catalytic system and an ethylene / butadiene copolymer obtained using said method, having butadiene units with 1,2-cyclohexane or 1,2- and 1,4-cyclohexane chains. The inventive complex has one of the following formulas (A and B), the complex is characterized in that in formula A, two molecules of ligands Cp<1> and Cp<2> are linked to the lanthanide Ln, such as Nd, said ligand molecules each consisting of a fluorenyl group, and, in formula B, a ligand molecule is linked to the lanthanide Ln, said ligand molecule consisting of two fluorenyl groups Cp<1> and Cp<2> which are interlinked by means of a bridge P having formula MR<1>R<2>, in which M is an element from column IVA, such as Si, and R<1> and R<2>, which may be identical or different, represent an alkyl group having between 1 and 20 carbon atoms.
Owner:MICHELIN & CO CIE GEN DES ESTAB MICHELIN +4
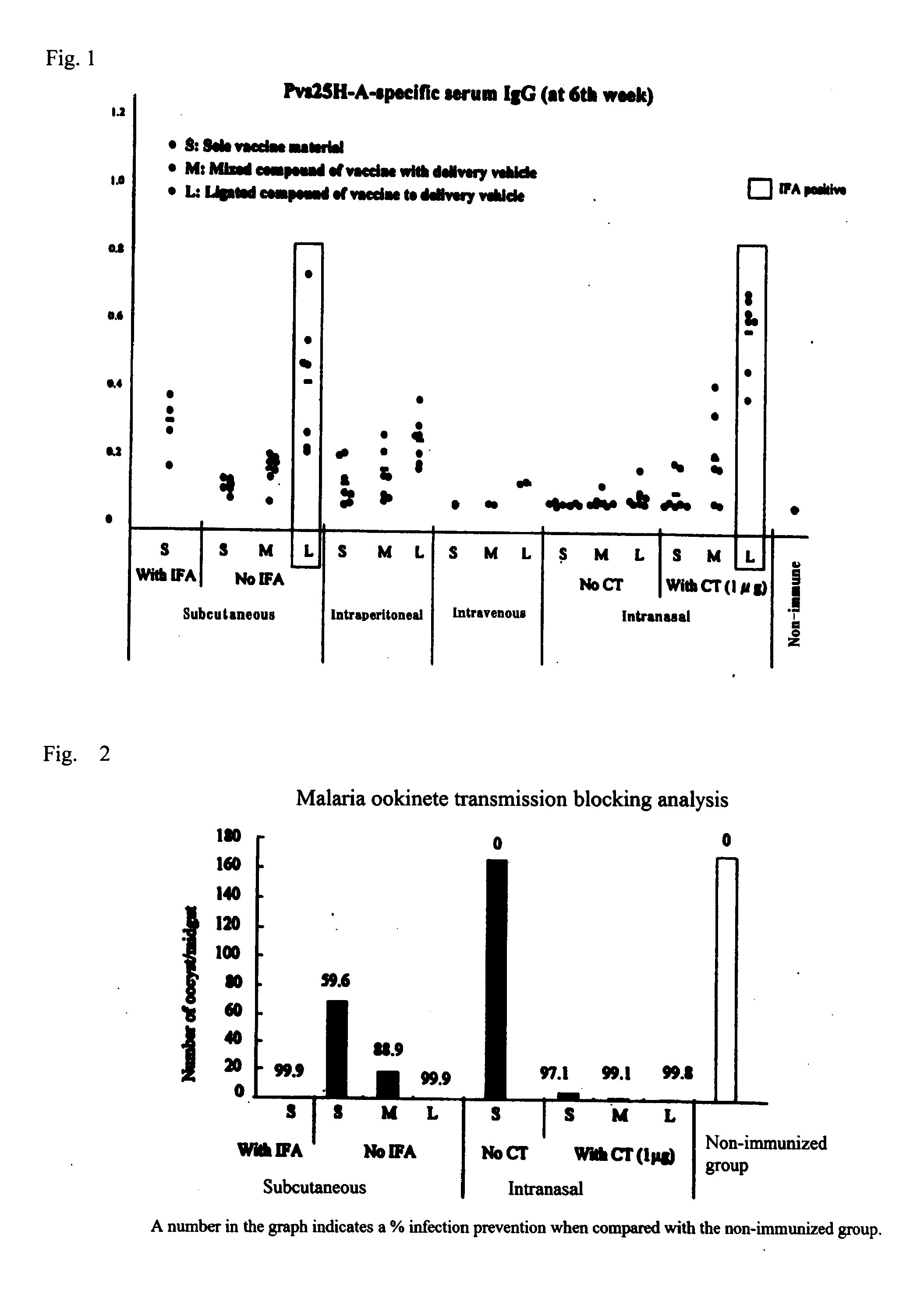
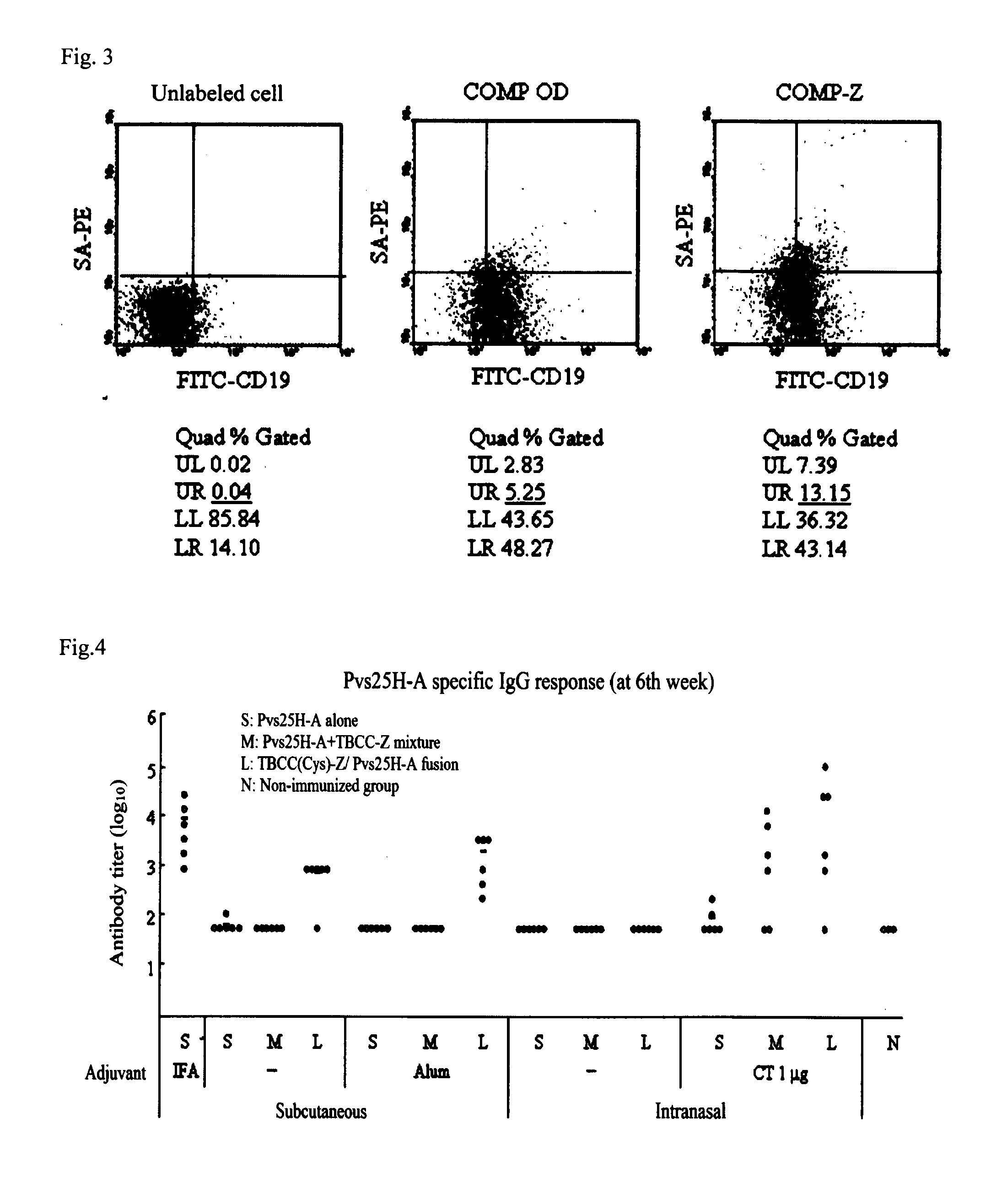
Drug transporter, and adjuvant and vaccine each utilizing same
InactiveUS20120100165A1Good effectImproving immunogenicityAntibody mimetics/scaffoldsAntiviralsAdjuvantImmunogenicity
An objective of the invention is to provide a drug delivery vehicle capable of allowing a vaccine or adjuvant to reach a target cell or tissue efficiently while being capable of improving the immunogenicity of the vaccine or capable of enhancing the immunostimulating effect of the adjuvant as well as a vaccine or adjuvant utilizing the same. Said drug delivery vehicle contains a multimeric protein having a coiled coil structure and a ligand molecule to a receptor of an immune cell.
Owner:UNIVERSITY OF THE RYUKYUS
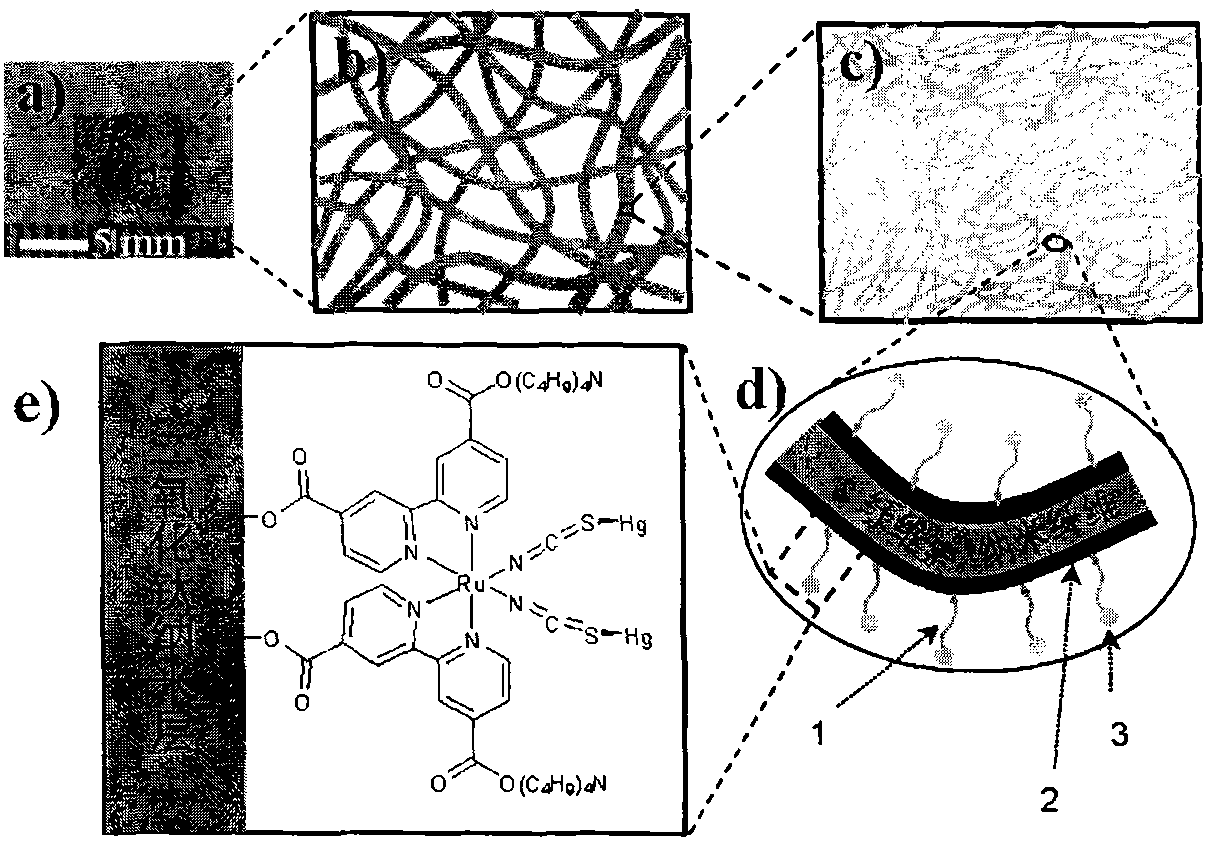
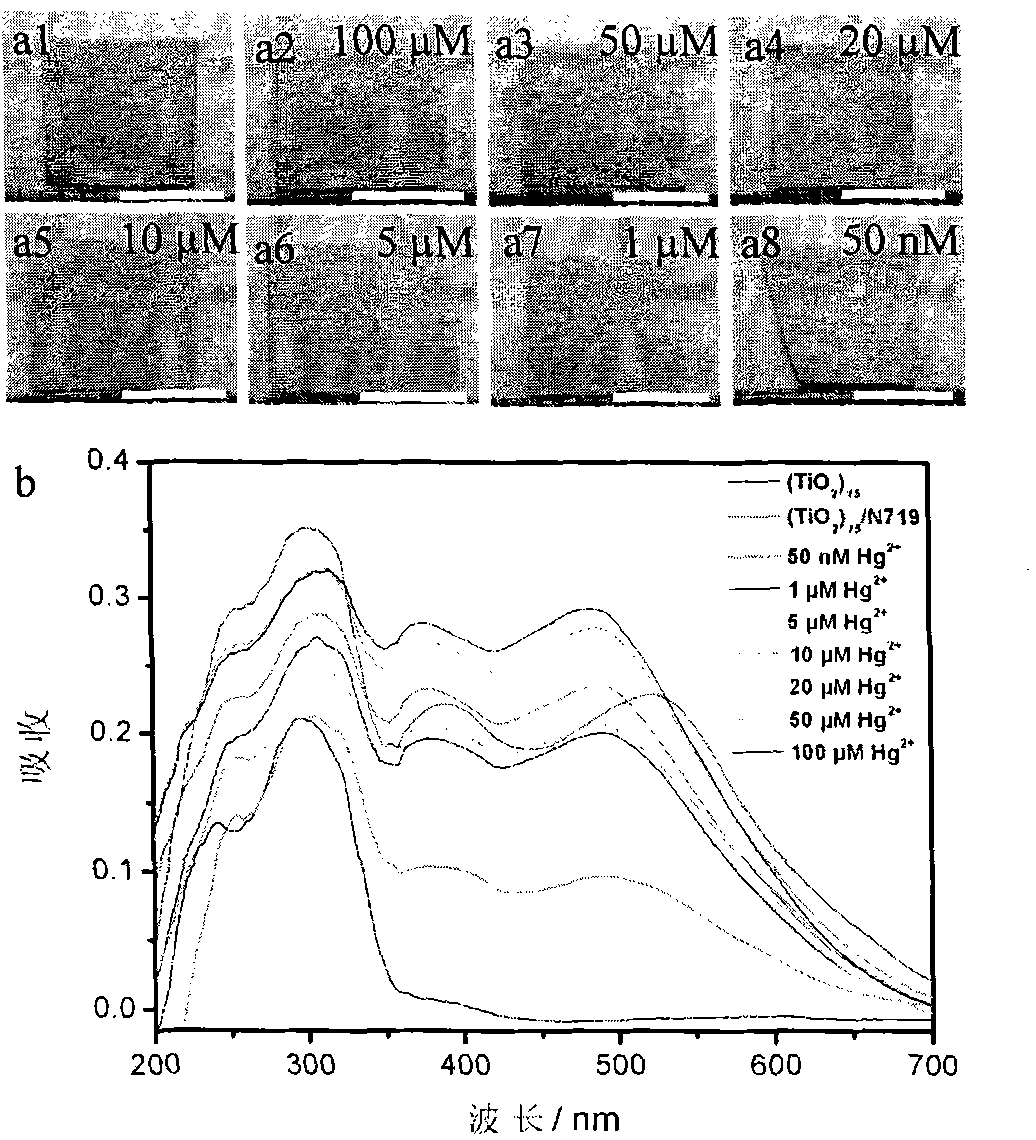
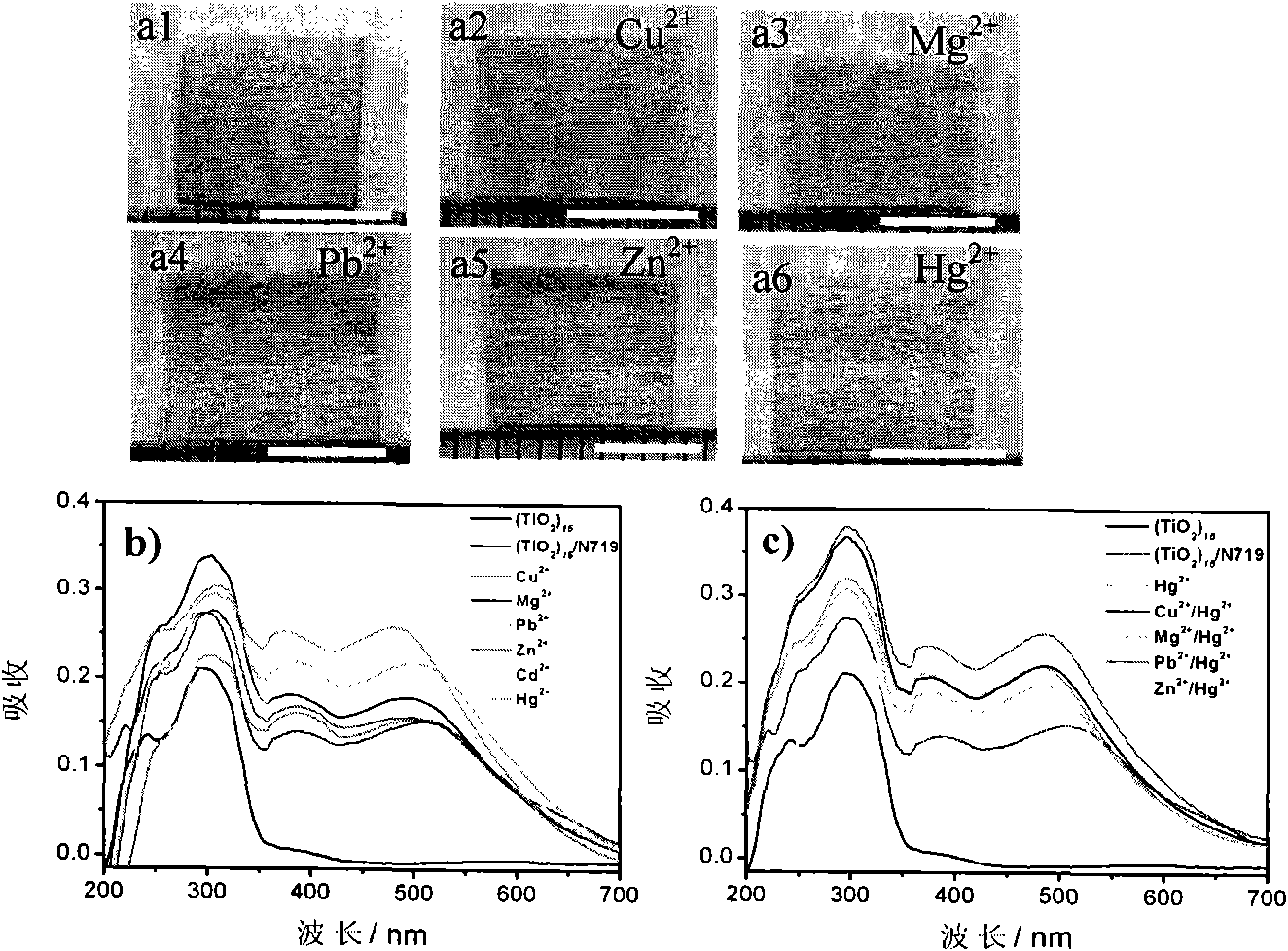
Preparation method of mercury ion colorimetric sensing cellulose material
InactiveCN101871164AHigh sensitivityEasy to prepareMaterial analysis by observing effect on chemical indicatorVegetal fibresCellulose fiberPotassium iodine
The invention discloses a preparation method of a mercury ion colorimetric sensing cellulose material, which comprises the following steps: taking a natural cellulose fiber material as a matrix, taking tetrabutyl titanate as a precursor and depositing a titanium dioxide film on the surface of a filter paper fiber by using a surface sol-gel method; then self-assembling and introducing a ruthenium dye N719 ligand molecule monolayer sensitive to mercury ions to obtain the mercury ion colorimetric sensing cellulose material; and detecting the mercury ions in the solution by simply infiltrating the material prepared by the invention in water solution of the mercury ions for a certain time and observing the change of the solution. In addition, the material prepared by the invention can also realize the recycling after treated by potassium iodide solution. The mercury ion colorimetric sensing cellulose material has excellent sensitivity and selectivity on the mercury ions and has the advantages of abundant sources of raw materials, low price, simple preparation method and capability of realizing simple, quick and economic detection on the mercury ions.
Owner:ZHEJIANG UNIV
Binding layer and method for its preparation and uses thereof
ActiveUS20070117199A1Enhance layeringSignificant comprehensive benefitsBioreactor/fermenter combinationsBiological substance pretreatmentsAnalyte moleculePolysaccharide
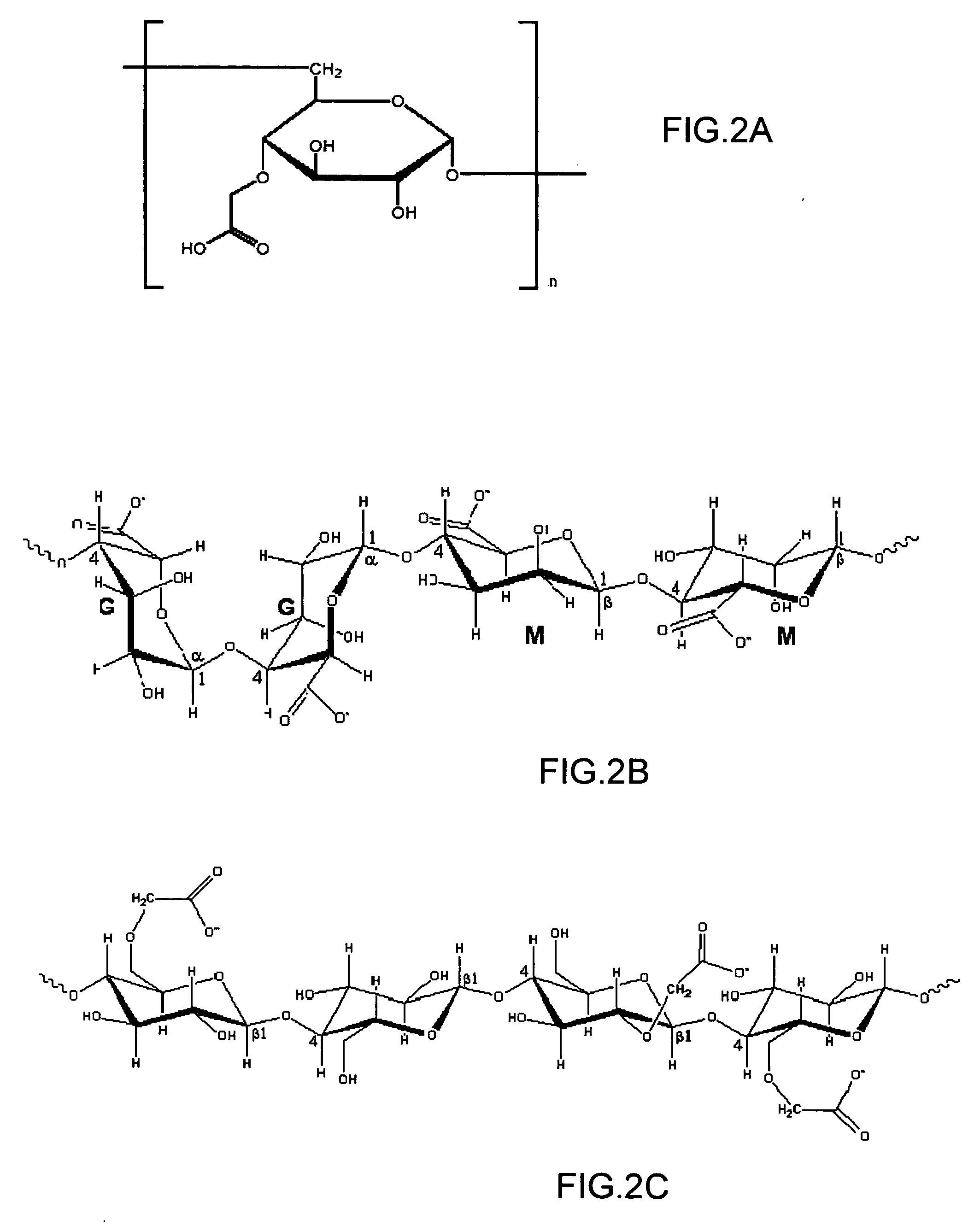
The invention provides a binding layer comprising a polysaccharide substituted by carboxylic groups or derivatives thereof exhibiting high performance in the binding of ligand molecules and in the interaction thereof with analyte molecules. A method for the preparation of the binding layer and for the assaying of various analyte molecules is also provided.
Owner:BIO RAD HAIFA
KIR and ligand genetic typing experimental method
InactiveCN108624665ASmall sample sizePromote amplificationMicrobiological testing/measurementDiseaseNatural Killer Cell Inhibitory Receptors
The invention discloses a KIR and ligand genetic typing experimental method. By the technology, the time and labor are saved, and meanwhile, DNA sample capacity is further saved. A multi-PCR technology is adopted, meanwhile, 36 pairs of primers are further combined according to the sizes of PCR products, and finally, amplified reaction is finished in 12 reaction holes. The KIR and ligand genetic typing experimental method is conveniently applied to amplification of 96 pore plates, conventional Taq enzyme is used, typing of all KIR genes and ligands thereof can be finished at a time under the same reaction conditions, and the practicality of the KIR and ligand genetic typing experimental method is greatly improved. Two pairs of primers are used for a KIR gene, the accuracy is improved, anda false negative result is avoided. The KIR gene can be combined to MHC-I type ligand molecules on the surfaces of targeting cells, inhibiting or activating signals are transmitted to regulate the activity of NK cells and T cells, and the KIR and ligand genetic typing experimental method plays an important regulation role in hematopoietic stem cell transplantation, feto-matemal tolerance, anti-infectious immunity, tumor immunity and autoimmune diseases. Therefore, KIR genetic typing facilitates understanding of influences of KIR to tumor immunity, hematopoietic stem cell transplantation and autoimmune diseases.
Owner:韩瑜
Methods for searching stable docking models of biopolymer-ligand molecule complex
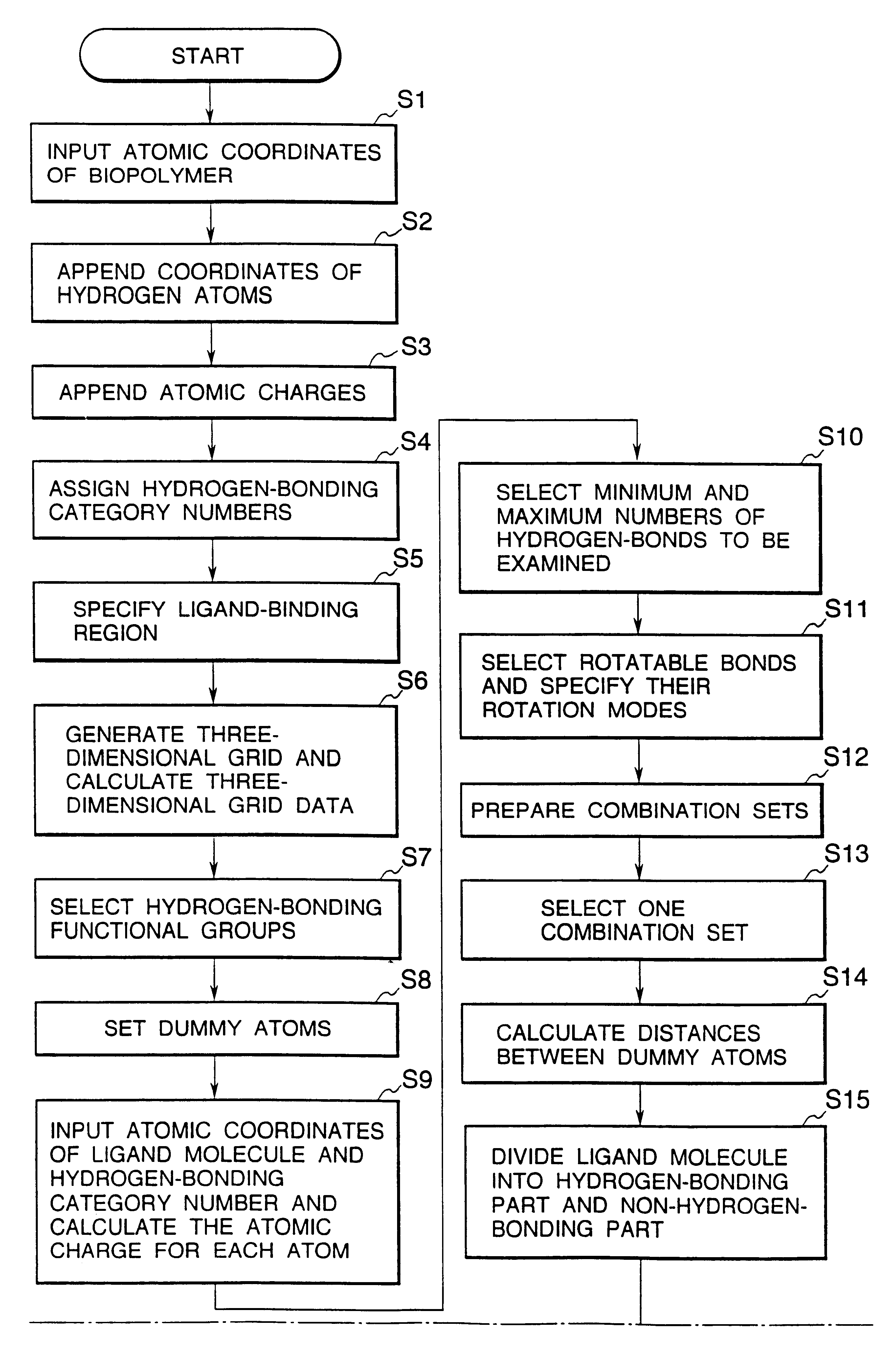
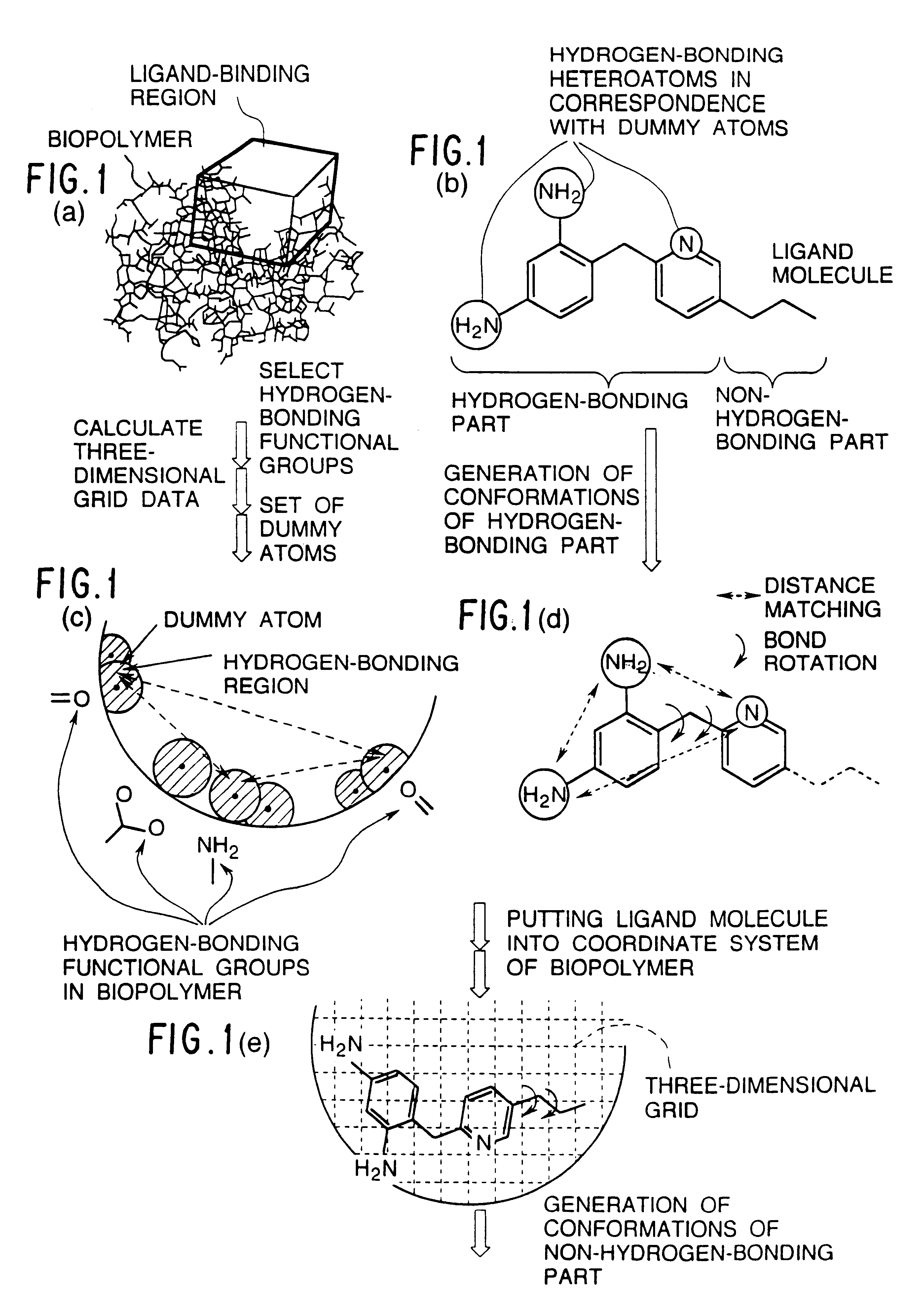
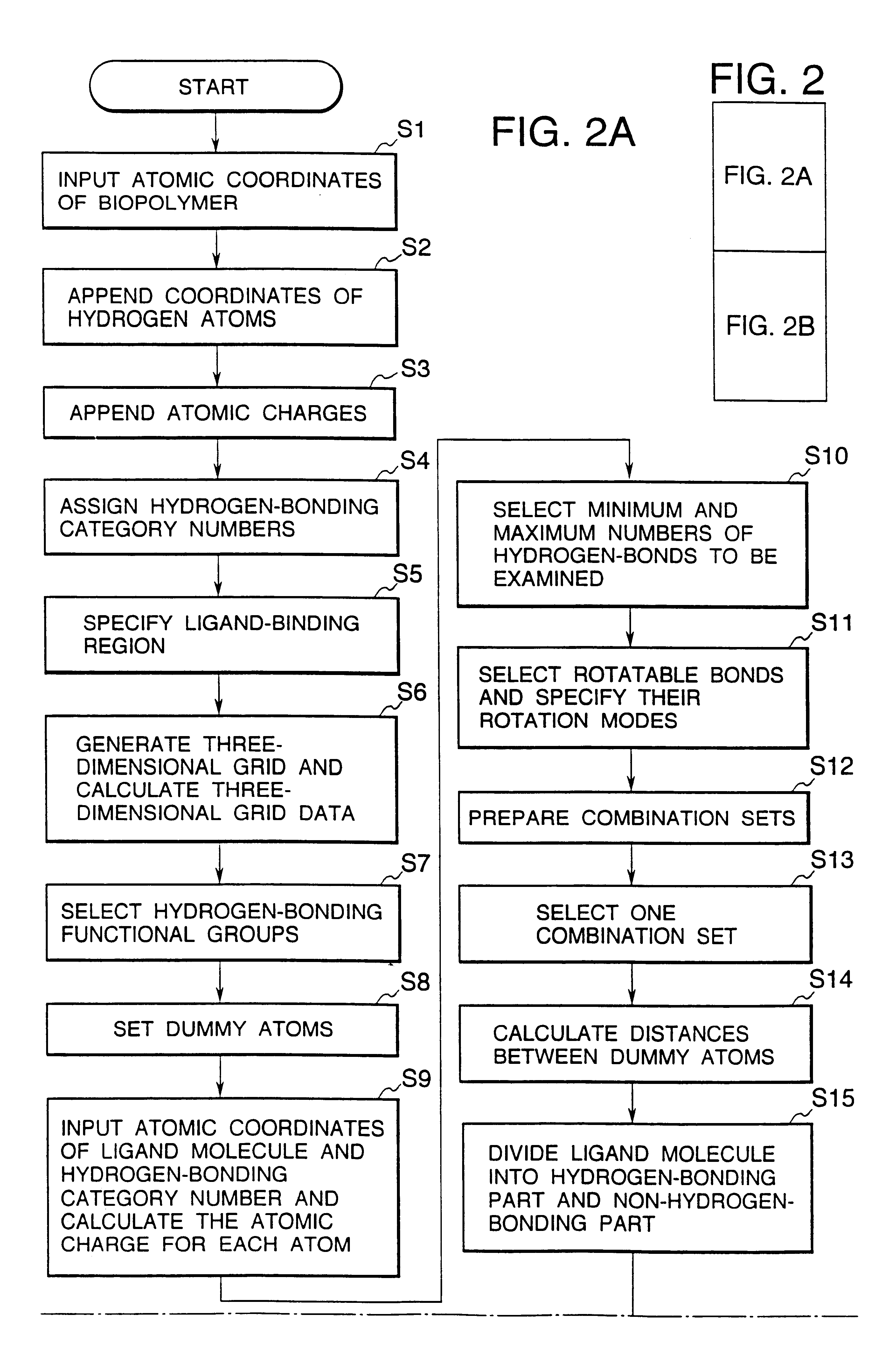
InactiveUS6308145B1Maximum flexibilityFlexible handlingAnalogue computers for chemical processesSpecial data processing applicationsHydrogenBiopolymer
Methods for searching stable docking models of biopolymer-ligand molecule complex, which comprise the steps of: (1) searching possible hydrogen-bond schemes between a biopolymer and a ligand molecule by preparing possible combination sets of hydrogen-bonding heteroatoms in the ligand molecule with dummy atoms located at the positions of heteroatoms that can be hydrogen-bond partners to hydrogen-bonding functional groups in the biopolymer; (2) estimating the possible the hydrogen-bond schemes between the biopolymer and ligand molecule and the possible conformations of a hydrogen-bonding part of the ligand molecule at the same time by comparing the distances between the dummy atoms with the distances between the hydrogen-bonding heteroatoms; and (3) obtaining the possible docking models of the biopolymer-ligand molecule complex by changing the coordinates of all atoms of the ligand molecule into the coordinate system of the biopolymer, according to the correspondences between the hydrogen-bonding heteroatoms in the ligand molecule and the dummy atoms in combination sets for each of the hydrogen-bond schemes and conformations obtained in the second step are provided. According to the methods of the present invention, the structures of stable biopolymer-ligand molecule complex can be searched efficiently and precisely in a short time.
Owner:INST OF MEDICINAL MOLECULAR DESIGN INC 50
Method for preparing monoatomic catalysts
ActiveCN109012732AAccurate and controllable spacingRegulated contentPhysical/chemical process catalystsHigh energyBenzaldehyde
The invention discloses a method for preparing monoatomic catalysts. The method comprises the following steps: step 1, preparation of a transition metal precursor: separately weighing and adding organic amine and substituted benzaldehyde into an acetonitrile solution to obtain an intermediate product, carrying out reduction, extraction and drying to obtain an organic ligand molecule product, separately dissolving organic ligand molecules and a transition metal salt in an acetonitrile solution, continuing mixing under stirring complete dissolving, and successively carrying out filtering, washing and drying so as to obtain a transition metal precursor product; step 2, mechanical ball milling: dispersing the transition metal precursor in carbon black with a high specific surface area by usinga high-energy mechanical ball milling process to obtain a monoatomic catalyst precursor; and step 3, carrying out high-temperature calcinations so as to obtain corresponding monoatomic catalysts. Theinvention realizes the preparation of transition metal monoatomic catalyst materials, has good expandability and reproducibility, and solves the problems of low metal content, few varieties and complicated preparation process in the prior art.
Owner:SUN YAT SEN UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com