Method for predicting binding free energy of protein and ligand based on progressive neural network
A neural network and neural network training technology, which is applied in the field of predicting the binding free energy of proteins and ligands based on progressive neural networks, can solve problems such as poor correlation of calculation results, low accuracy, and large calculation time, and achieve The effect of avoiding over-fitting problems, good generalization ability, and speeding up calculation speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
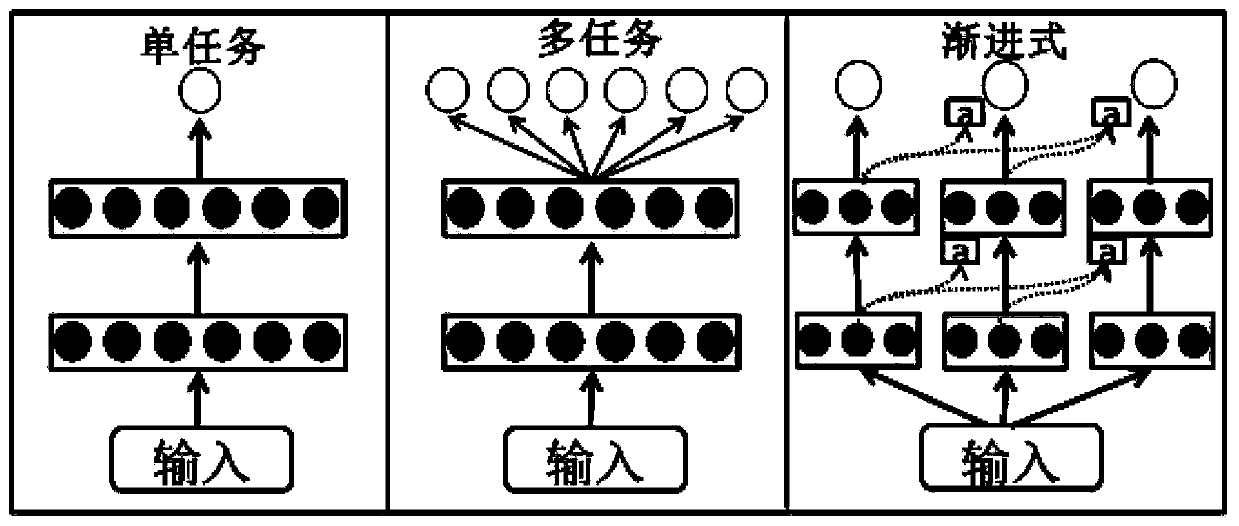
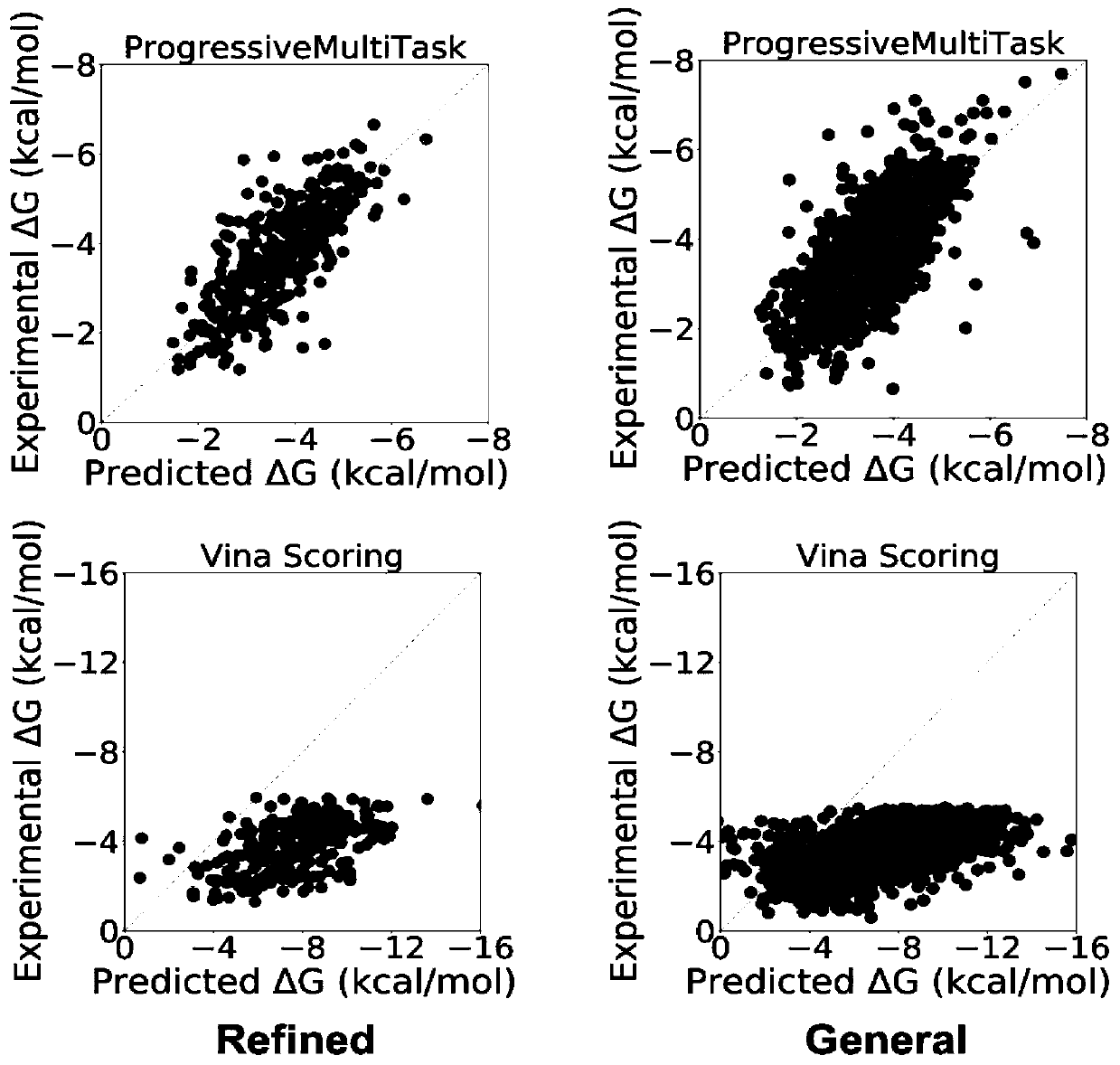
[0036] Such as Figure 1-Figure 4 A method for predicting protein-ligand binding free energy based on a progressive neural network is shown, comprising the following steps:
[0037] Step 1: Establish a database server, a structural information processing server and a neural network training server, and the database server, structural information processing server and neural network training server all communicate with each other through the Internet;
[0038] Based on the crystal structure data in the PDB database and PDBbind database, after data preprocessing, the PDBLig database is established in the local database server to store the pdb files of proteins and ligand molecules.
[0039] The process of data preprocessing includes downloading data from PDB and PDBbind databases, deleting files with incomplete structures or no ligand molecules or no experimental binding constants.
[0040] In this embodiment, the analysis accuracy of each pdb file is classified in the PDBbind da...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com