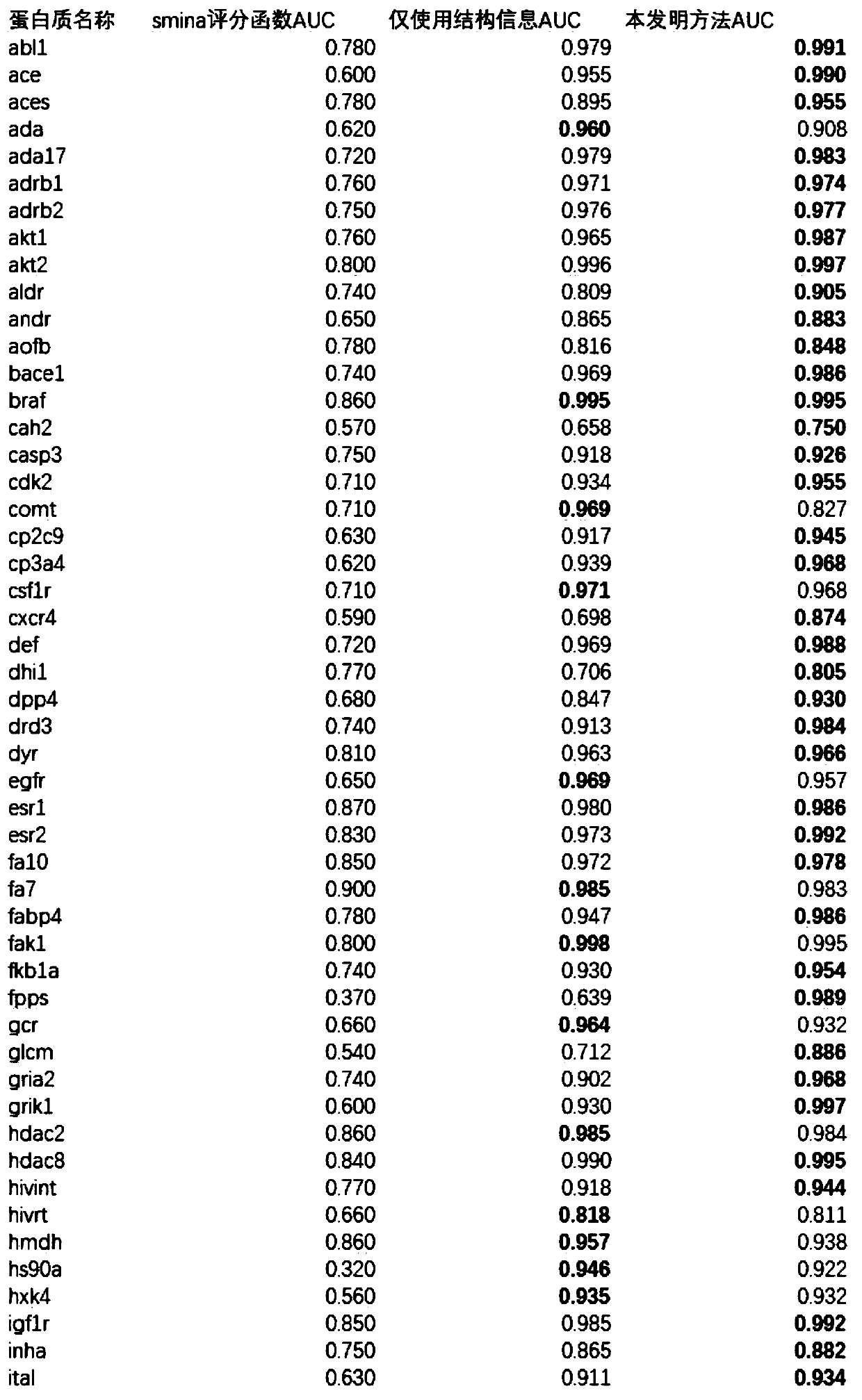
Patents
Literature
172 results about "Molecule docking" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
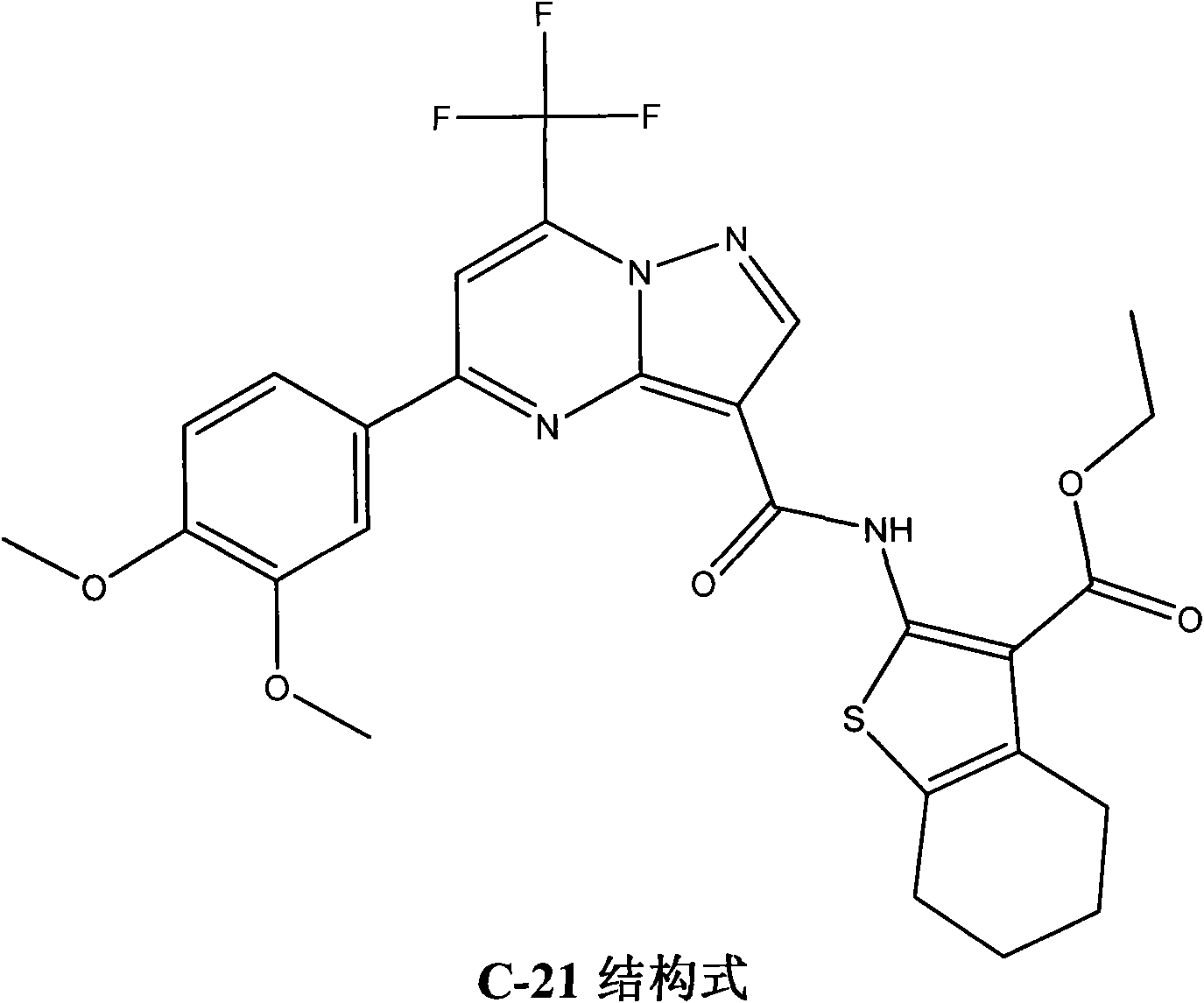
Application of N-(thiofuran-2) pyrazolo (1, 5-a) pyridine-3-formanides compounds for preparing antineoplastic
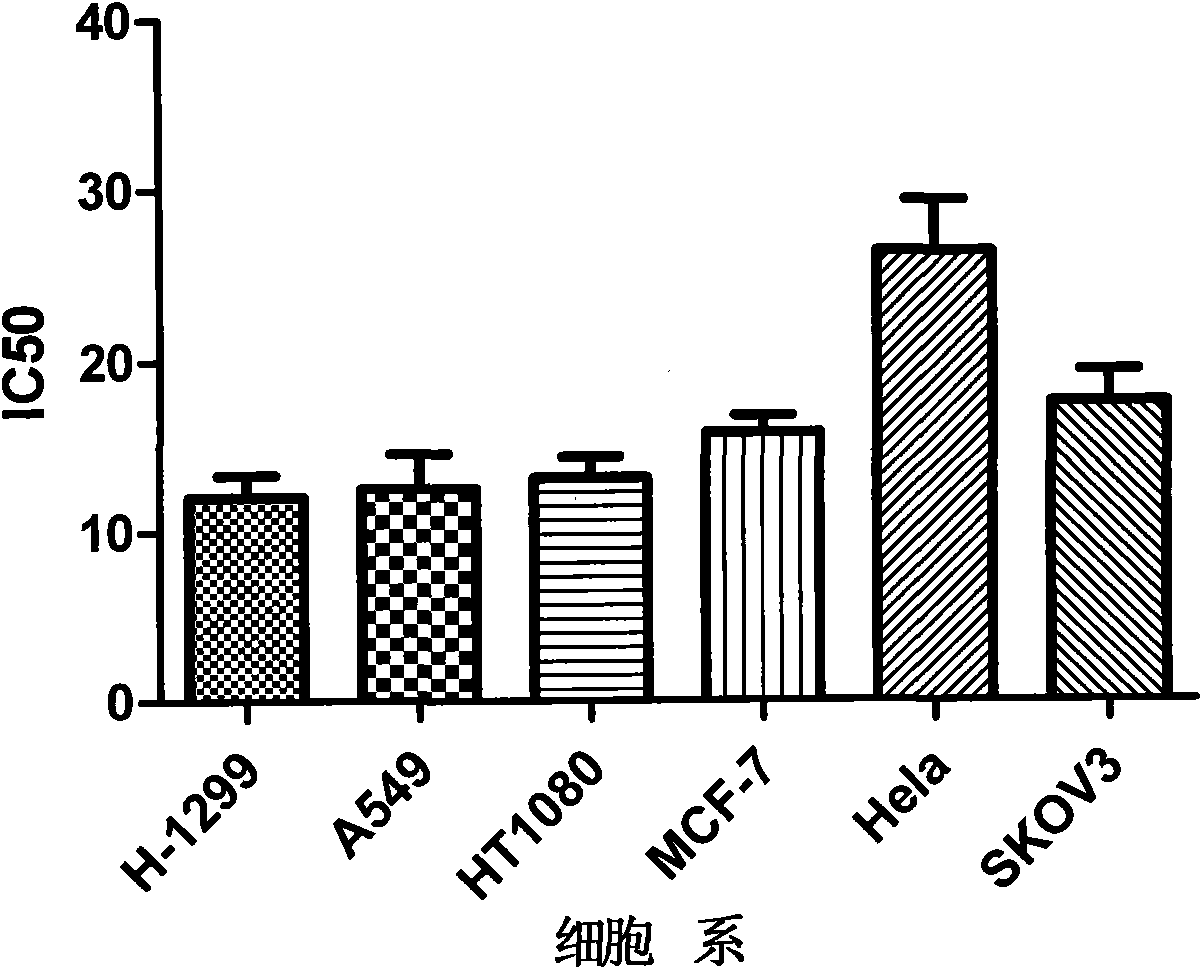
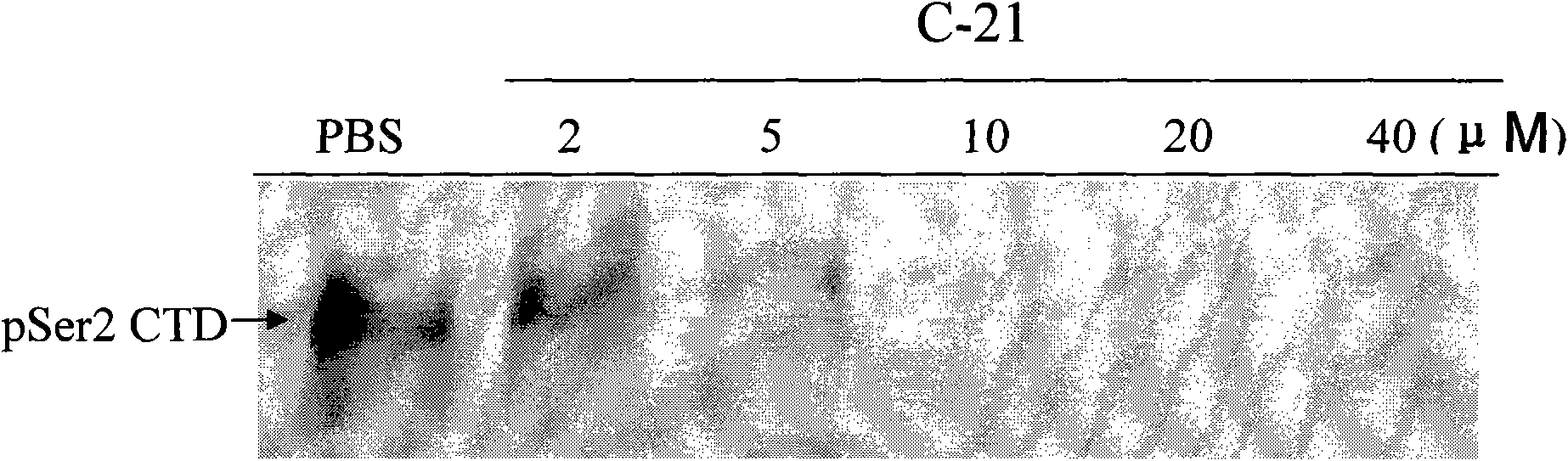
The invention searches the novel micromolecule inhibitor pyrazolo (1, 5-a) miazines compounds of cyclin-dependent kinase CDK9 (cyclin-dependent kinase) through the virtual screening of a computer, biometrically measures activity thereof, and validates interaction mechanism. The invention specifically comprises the following steps: the three-dimensional crystal conformation of the cyclin-dependent kinase family member CDK9 is obtained in a way of homology modeling; and micromolecule three-dimensional database is screened with DOCK (molecular docking). The invention uses a MTT tumor cell growth inhibition test to biometrically measures the activity of the selected compounds, researches the selected compounds pyrazolo (1, 5-a) miazines with high activety in a way of molecular mechanism, validates the inhibiting effect of the compounds to the activity of CDK9 kinase, and clarifies the interaction mechanism of the compounds for inhibiting the external activity and the molecule of various malignancies such as lung cancer, osteosarcoma, oophoroma, cervical carcinoma, breast cancer, etc.
Owner:INST OF HEMATOLOGY & BLOOD DISEASES HOSPITAL CHINESE ACADEMY OF MEDICAL SCI & PEKING UNION MEDICAL COLLEGE
Method for predicting binding free energy of protein and ligand based on progressive neural network
ActiveCN110910951APrevent overfittingImprove generalization abilityProteomicsGenomicsAlgorithmTest set
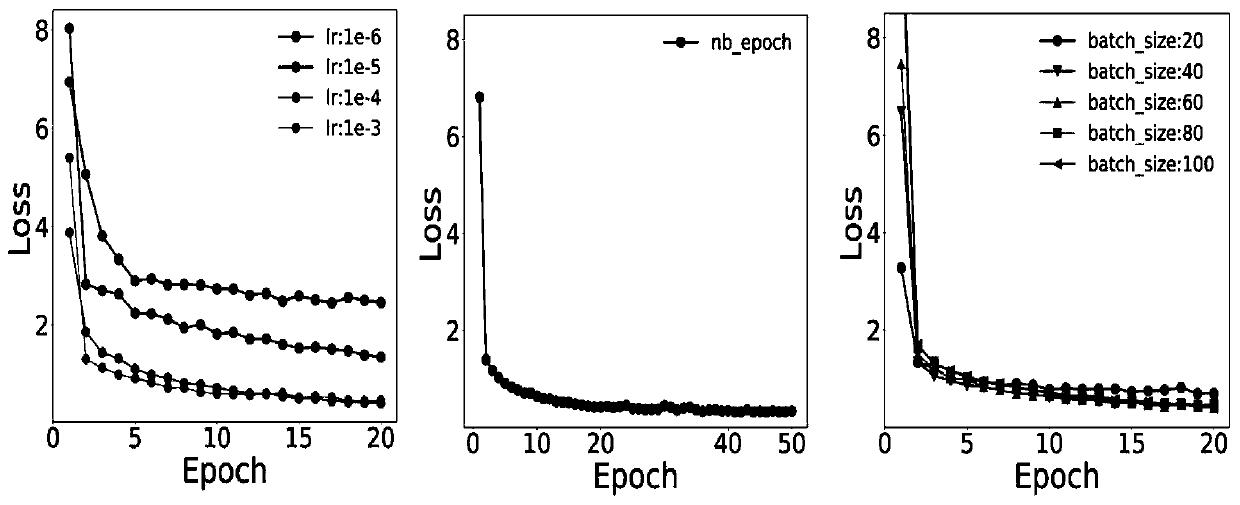
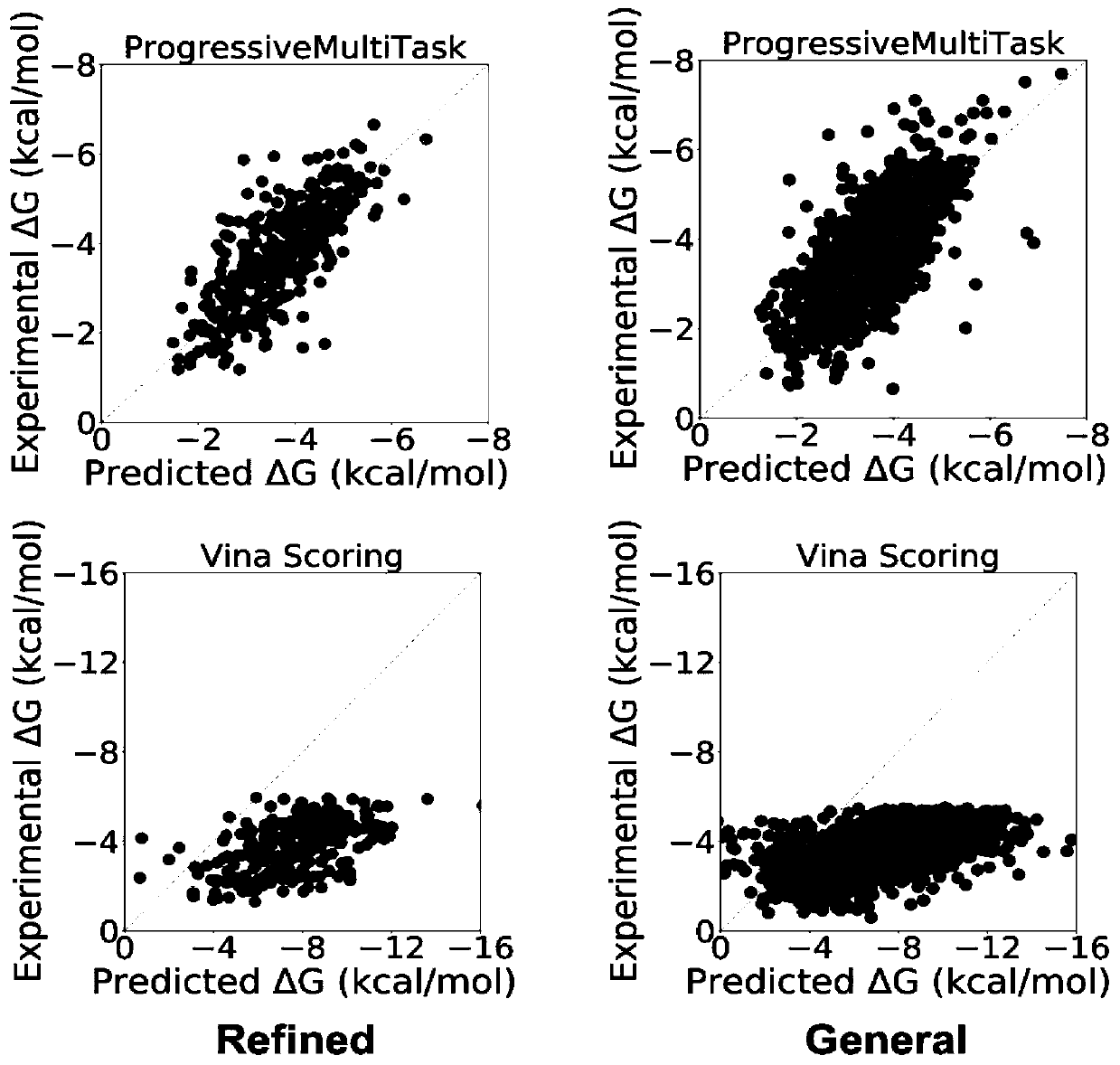
The invention discloses a method for predicting the binding free energy of protein and ligand based on a progressive neural network, and belongs to the technical field of computer-aided drug design. The method comprises the steps: obtaining a pdb file from a PDBbind database, establishing local database, acquiring an amino acid molecule within 4.5 angstroms in the protein binding pocket by takingthe ligand molecule as a center, performing extended connectivity fingerprint calculation, carrying out SPLIF fingerprint calculation, searching for the number of salt bridges and hydrogen bonds between protein and ligand molecules, converting the structural information of the protein and the ligand into a one-dimensional tensor, and establishing a training set, a verification set and a test set;training the progressive neural network by using the training set; optimizing and searching hyper-parameters for prediction; through comparison with a molecular docking result, obtaining a higher Pearson correlation coefficient. According to the invention, the technical problem of how to convert a three-dimensional structure of protein and ligand molecules into tensors which are easy to calculateby a computer and input the tensors into the progressive neural network for training and optimization is solved, and the calculation rate and the prediction accuracy are greatly improved.
Owner:JIANGSU UNIV OF TECH
Virtual drug screening method and device, computing equipment and storage medium
ActiveCN111462833AFocus on physical and chemical propertiesFocus on dynamic featuresChemical property predictionMolecular designProtein targetAlgorithm
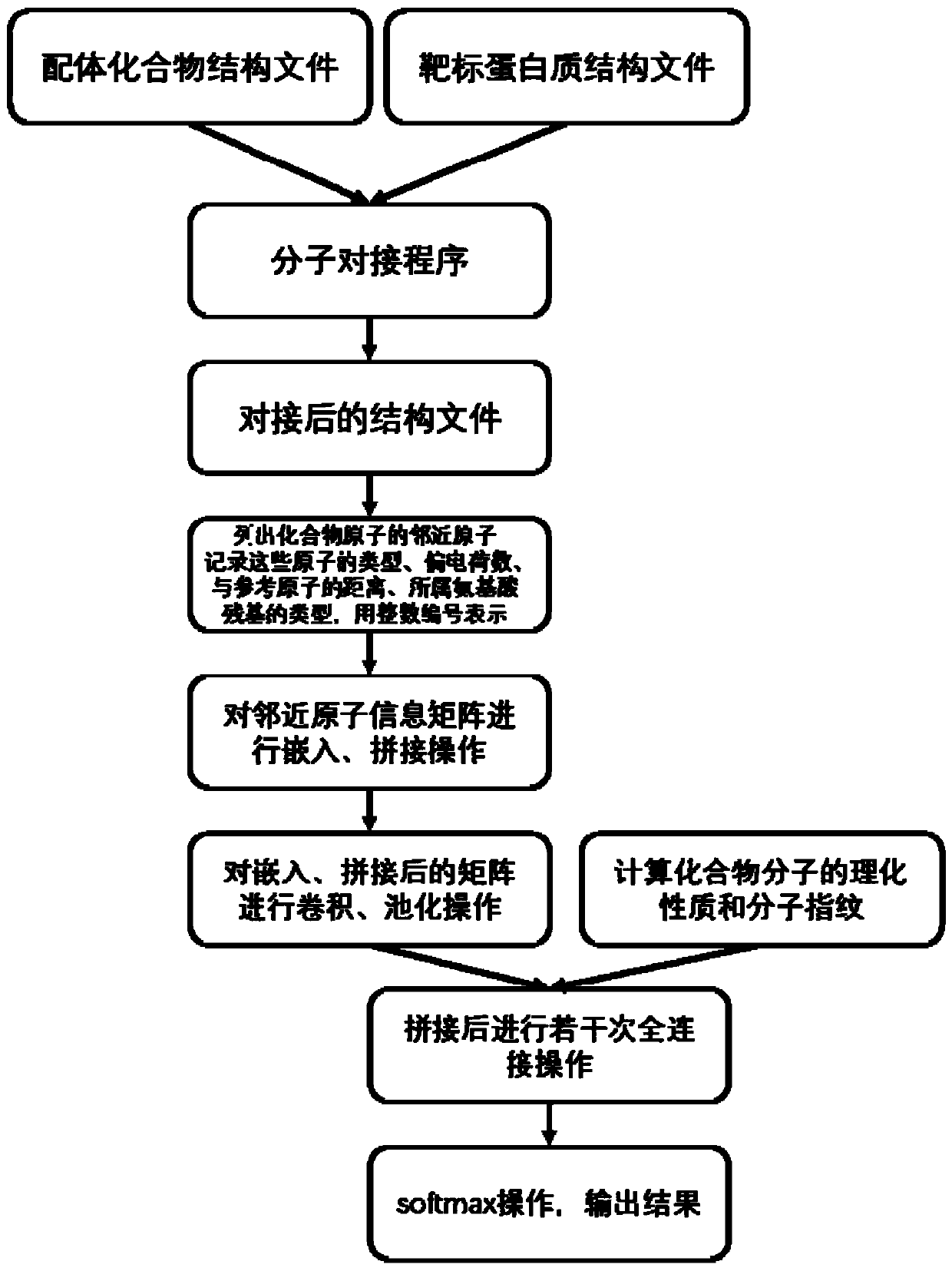
The invention discloses a virtual drug screening method and device, computing equipment and a storage medium. The method comprises the following steps: carrying out molecular docking on a ligand compound and a target protein; taking each atom contained in the docked compound molecules as a reference atom, determining a compound adjacent atom and a protein adjacent atom of each reference atom, recording corresponding predetermined structure information, and mapping the predetermined structure information into a structure information matrix group; performing embedding operation on the structureinformation matrix group by using a neural network, and obtaining a representation matrix of a compound-protein complex from the embedded structure information matrix group; carrying out convolution,bias and pooling on the representation matrix to obtain a structure vector; and splicing the structure vector with a physicochemical property vector representing the physicochemical property and the molecular fingerprint of the compound, and performing full-connection operation after neural network weighting and biasing to obtain a two-dimensional vector representing the inactivity and activity ofthe compound to the target protein so as to perform drug screening.
Owner:深圳智药信息科技有限公司
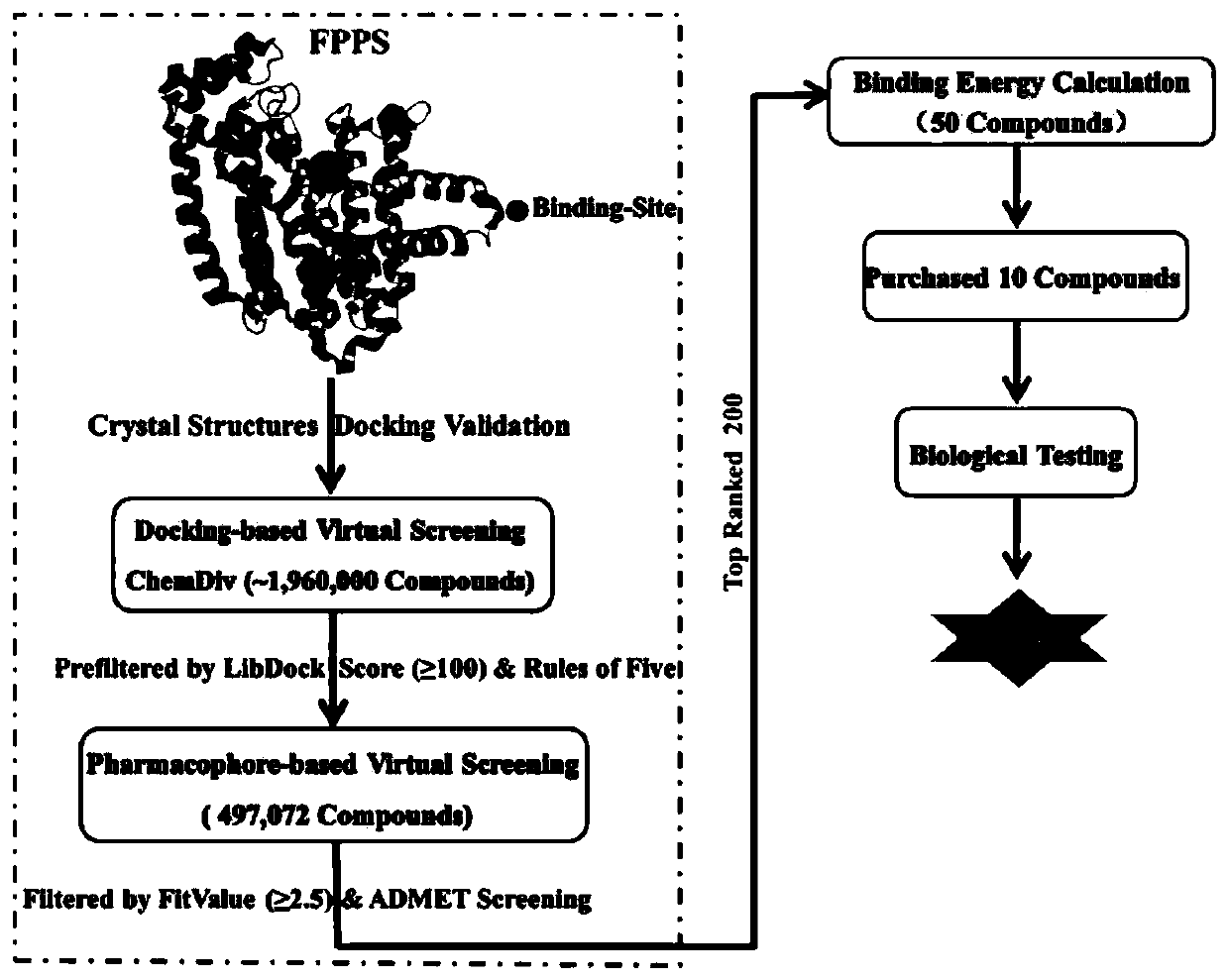
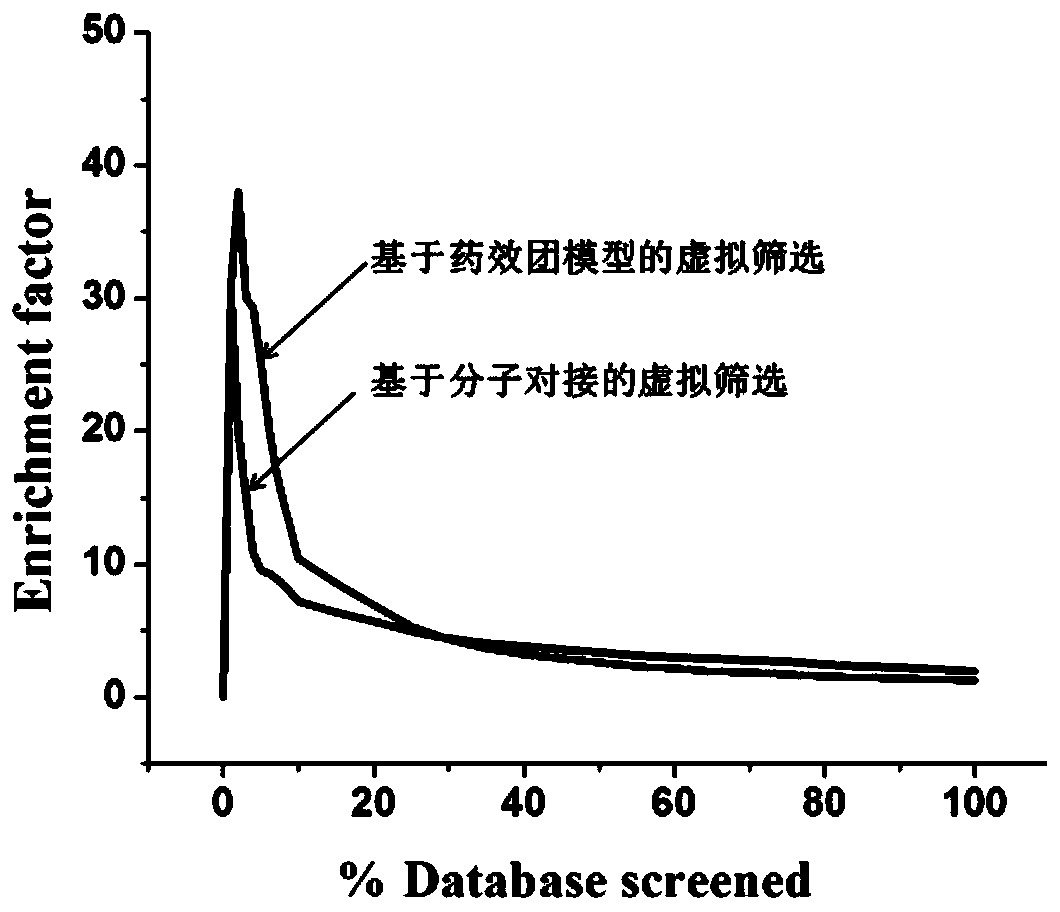
Screening method and application of farnesyl pyrophosphate synthase FPPS inhibitor
ActiveCN110289054AReduce blindnessImprove hit rateChemical property predictionMolecular designData setScreening method
The invention provides a screening method of a farnesyl pyrophosphate synthase (FPPS) inhibitor. The screening method comprises the following steps: (1) constructing an FPPS template; (2) preparing a data set; 3) performing virtual screening based on molecular docking; 4) performing virtual screening based on the pharmacophore model; 5) calculating binding energy; 6) integrating the docking scores in the steps 3), 4) and 5), the pharmacophore model FitValue value and the binding energy to obtain the FPPS inhibitor; and 7) carrying out FPPS enzyme activity inhibition experiment on the FPPS inhibitor obtained by virtual screening, and determining the FPPS inhibitor with the structure shown in the formula I. The method gives full play to the advantages of theoretical screening, reduces the blindness of drug synthesis, effectively improves the hit rate of positive drugs, saves manpower, material resources and financial resources, and realizes the new use value of old drugs. The invention further provides application of the FPPS inhibitor obtained through screening in preparation of drugs for treating colon cancer.
Owner:JIANGSU INST OF NUCLEAR MEDICINE
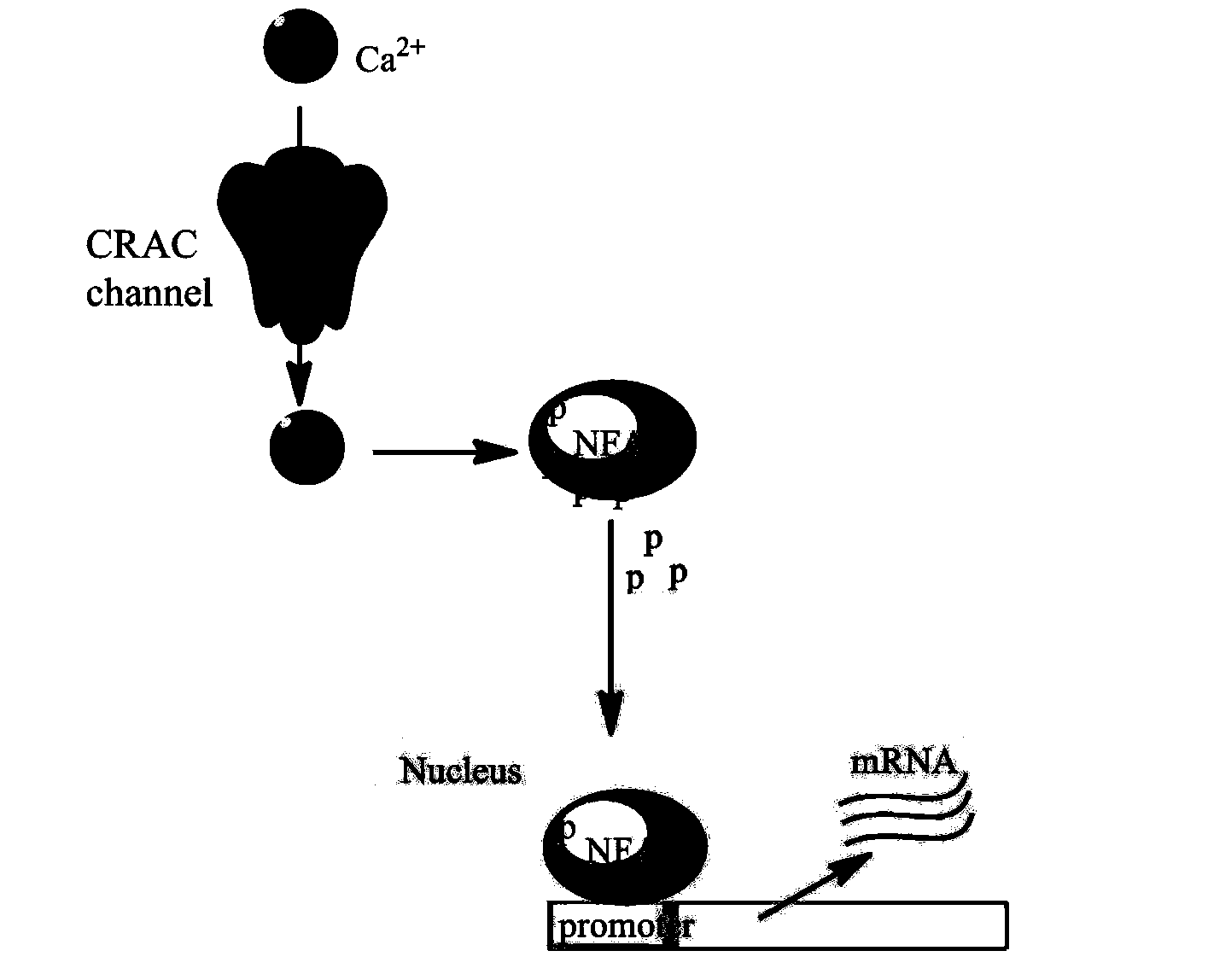
Virtual screening method for anti-inflammation and anti-rejection drugs taking CRAC channels as targets
ActiveCN104298891AImprove hit rateShort cycleMolecular designSpecial data processing applicationsVirtual screeningPharmacophore
The invention discloses a virtual screening method for anti-inflammation and anti-rejection drugs taking CRAC channels as targets. The virtual screening method comprises the following steps that (1), CRAC channel protein structural data are obtained to construct a homology model; (2), an active center is determined, and an activity bag is set; (3), according to the activity bag, molecular docking software is utilized for carrying out docking marking on compounds; (4), the compounds with the CRAC channel blocking activity are determined preliminarily; (5), activity screening is carried out on the compounds, and leading drugs with the CRAC channel blocking activity are obtained; (6), a pharmacophore model is constructed, the small-molecule compounds on which molecule docking is not carried out or the small-molecule compounds on which docking is carried out are matched and compared for guiding screening of the compounds or structure optimizing of the leading compounds. According to the virtual screening method for the drugs, the number of the compounds on which activity testing is carried out is reduced, the cost is saved, the screening efficiency is improved, and the virtual screening method can be used for further developing the novel anti-inflammation and anti-rejection drugs.
Owner:SHANDONG UNIV
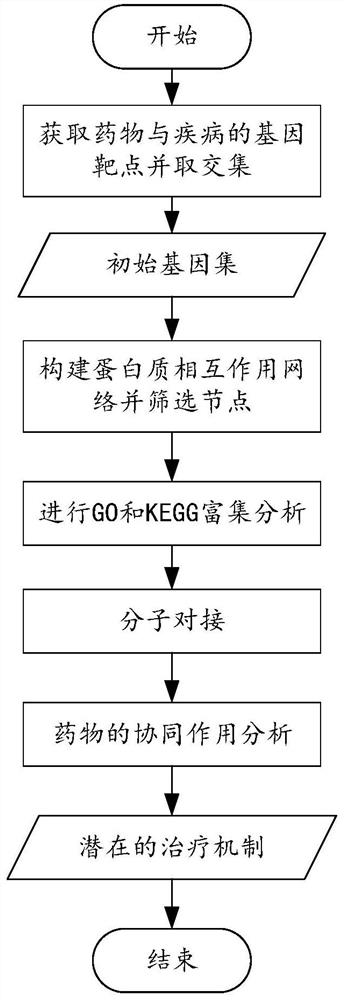
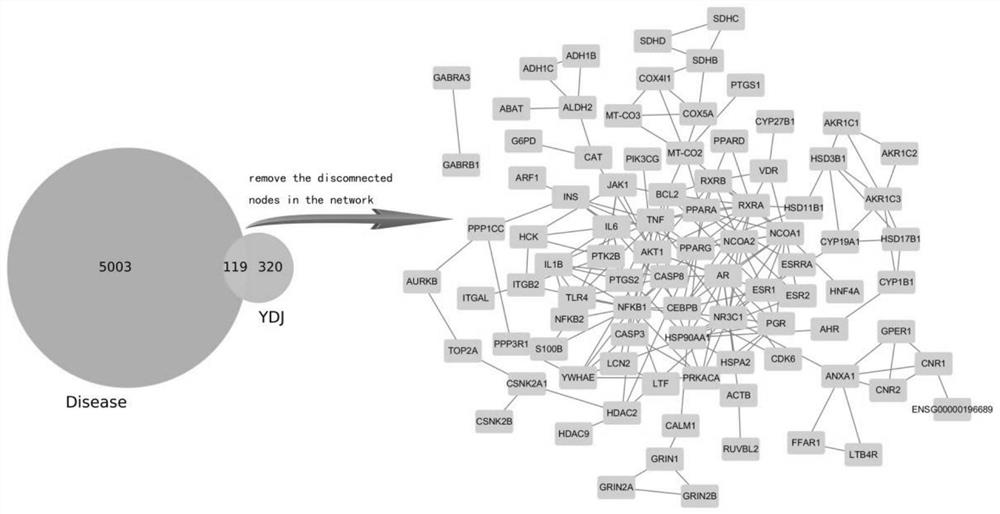
Method, system and device for establishing traditional Chinese medicine action mechanism model based on network pharmacology
ActiveCN111613297APredict interactionMolecular designAlternative medicinesPharmacometricsNetwork pharmacology
The invention provides a method, a system and a device for establishing a traditional Chinese medicine action mechanism model based on network pharmacology. The method comprises the following steps: acquiring gene targets corresponding to various components of the traditional Chinese medicine; acquiring a target gene target; taking an intersection of the gene targets corresponding to the components of the traditional Chinese medicine and the target gene targets to obtain a gene set; constructing a protein interaction network for the gene set; analyzing nodes in the protein interaction networkto obtain key gene nodes; performing GO analysis and KEGG analysis on the key gene nodes for further gene screening; in the screening result, conducting molecular docking on the traditional Chinese medicine components and protein corresponding to the core gene, verifying the binding capacity, and obtaining the target traditional Chinese medicine components. According to the scheme, efficient drugcomponent analysis can be assisted, the analysis process is simplified, the complexity of an analysis algorithm is reduced, convenience and rapidness are achieved, and the accuracy is high.
Owner:SICHUAN UNIV
Epitope polypeptide combination capable of inducing immunity and application thereof
PendingCN113372417AVerify immune response propertiesHighly conservativeSsRNA viruses positive-senseViral antigen ingredientsAntigen epitopeCtl epitope
The invention discloses an epitope polypeptide combination capable of inducing immunity and application thereof, belongs to the technical field of biology, and aims to carry out molecular design of related vaccines by utilizing immunoinformatics on the basis of epitope analysis optimization. Based on a structural antigen epitope vaccine design strategy, a B cell epitope, a Th epitope and a CTL epitope on a new coronavirus S protein are determined through immunoinformatics to induce a main neutralizing antibody, activate cellular immune response and induce body fluid and cellular immune balance. The epitope vaccine is designed through connection of a molecular adjuvant and candidate antigen epitopes, antigenicity, physicochemical properties, protein secondary structure and tertiary structure modeling of the epitope vaccine are analyzed, vaccine conformation B cell epitopes are analyzed by means of a structural biological tool, and immune response characteristics of the vaccine are verified through molecular docking with TLR4 and immune response simulation stimulation. Information analysis results show that the designed candidate epitope combination has well balanced humoral immune and cellular immune response capabilities.
Owner:SHANTOU UNIV MEDICAL COLLEGE
Amine transaminase AcATA mutant and application thereof in preparation of sitagliptin intermediate
ActiveCN111549008AIncrease enzyme activityImprove conversion rateBacteriaTransferasesKetonePhenylalanine
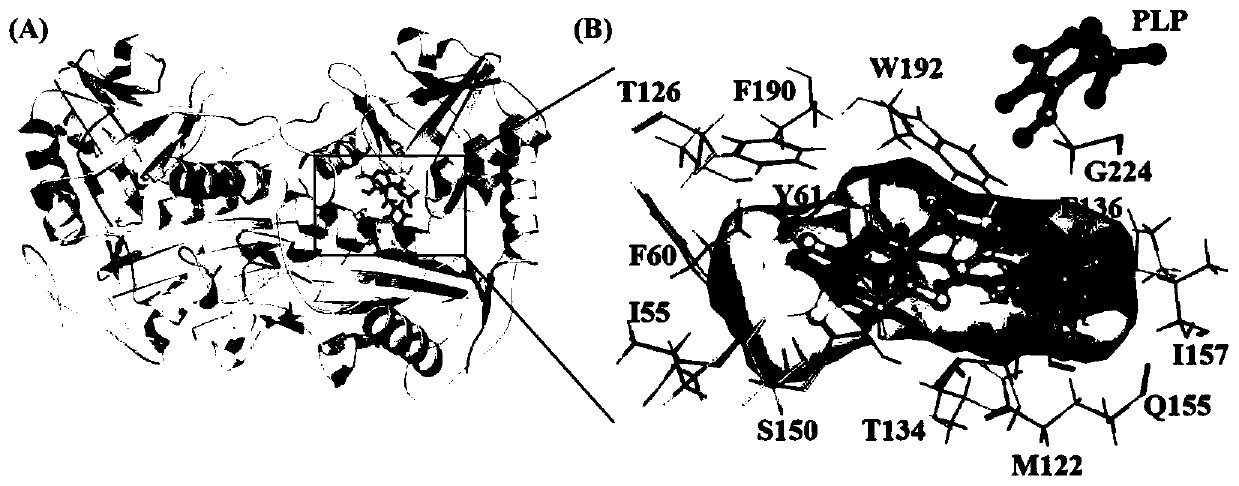
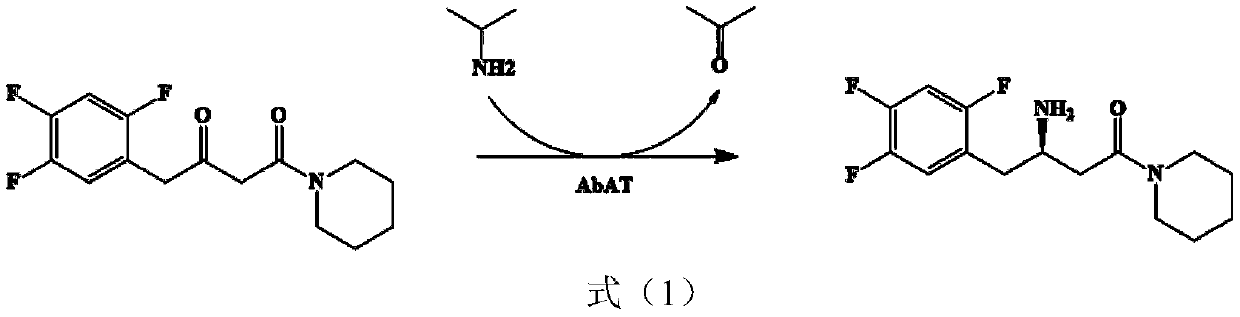
The invention discloses an amine transaminase AcATA mutant and application of the amine transaminase AcATA mutant in preparation of a sitagliptin intermediate. The amine transaminase AcATA mutant is obtained by single mutation at the 122th site of an amino acid sequence shown in SEQ ID No.2. The amino acid sequence of the amine transaminase AcATA mutant is shown in SEQ ID No.2. The amino acid sequence of the amine transaminase AcATA mutant is shown in the description, wherein methionine at the 122 position is mutated into histidine, valine or phenylalanine. According to the invention, sites possibly influencing the catalytic activity are obtained through molecular docking, homologous modeling and other methods, and site-specific mutagenesis is carried out, so that the finally obtained aminotransferase AcATA mutant has high enzyme activity, and the enzyme activity is higher than 460U / g and is more than 4 times of that of a wild type; the catalyst can efficiently catalyze the sitagliptin intermediate precursor ketone 1-piperidine-4-(2, 4, 5-trifluorophenyl)-1, 3-dibutanone to synthesize the sitagliptin intermediate (R)-3-amino-1-piperidine-4-(2, 4, 5-trifluorophenyl)-1-butanone, and the 24-hour conversion rate is up to 90%.
Owner:ZHEJIANG UNIV OF TECH +2
Structure prediction method of voltage-gating sodium ion channel
ActiveCN107247885AStructural prediction methods are reliableReliable methodMolecular designSequence analysisMacromolecular dockingVoltage
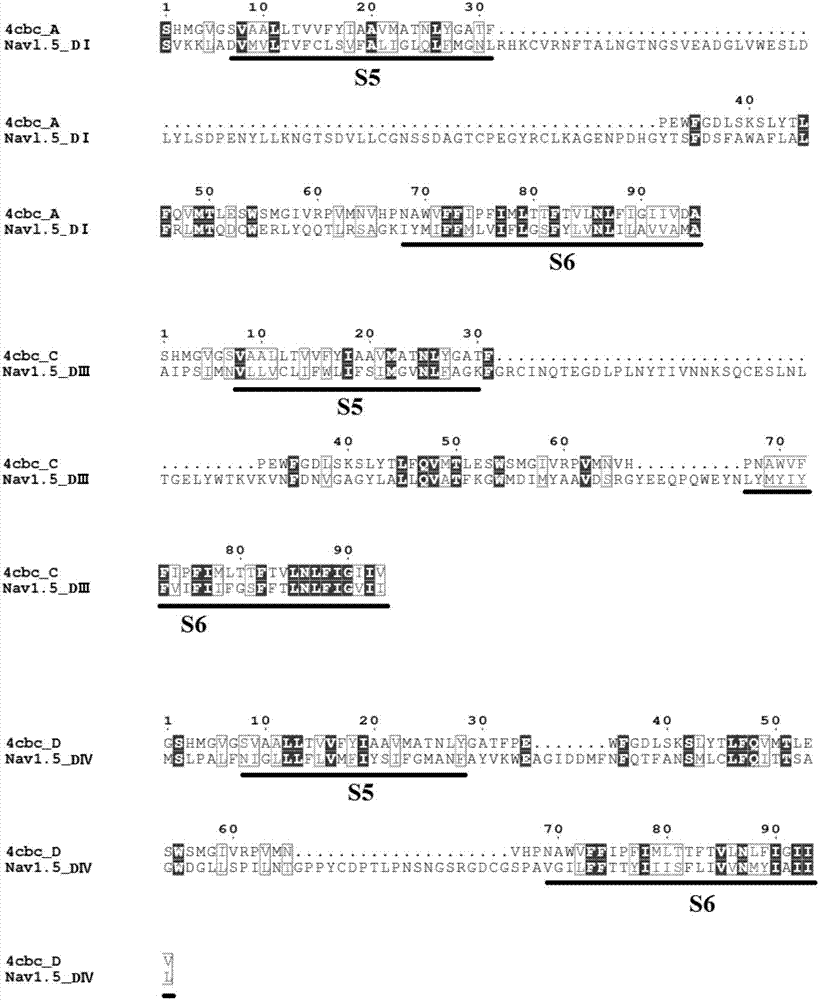
The invention discloses a structure prediction method of a voltage-gating sodium ion channel, and particularly relates to a structure modeling method of constructing the eukaryotic voltage-gating sodium ion channel Nav1.5 pore structural domain open state based on a template. The method includes the steps that a rosetta membrane protein homology modeling method is used for obtaining a germ sodium ion channel template of a crystal tripolar structure, and four subunit structures of the Nav1.5 pore structural domain are constructed and composed preliminarily; sorting is carried out according to the grading condition of all construction models, the structure with the maximum score is selected as the initial structure of all the structural domains; the four constructed subunit structures are compared to four 4CBC subunits, and the assembled overall structure is optimized; based on structure data and test data of existing local anesthetics drugs, molecular docking is carried out on the optimized structures, screening and evaluating strategies are set according to the existing experiment data to determine the reliability of model construction.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
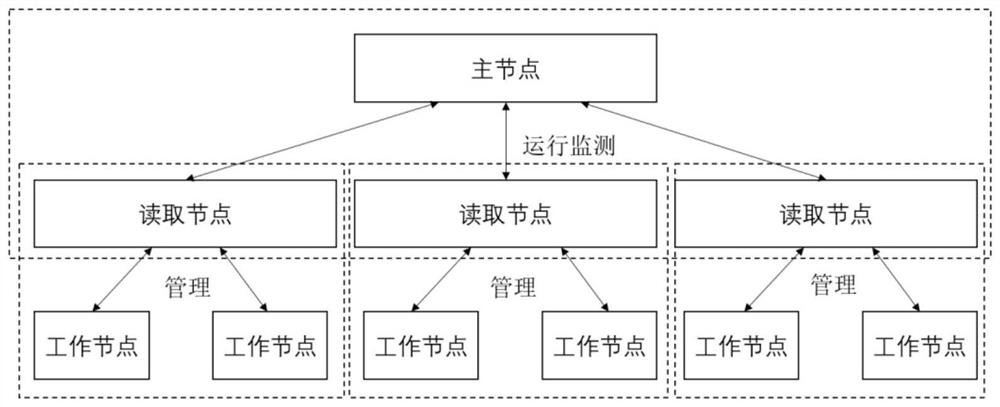
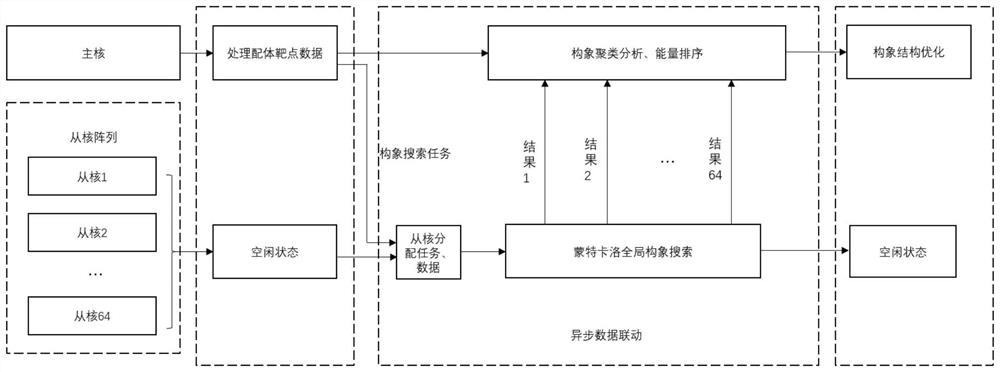
Ultra-large-scale marine natural product molecular docking method based on heterogeneous many-core architecture
ActiveCN114743613AReduce I/O pressureImprove stabilityMolecular designClimate change adaptationConcurrent computationVirtual screening
The invention relates to a super-large-scale marine natural product molecular docking method based on a heterogeneous many-core architecture, and belongs to the technical field of drug screening, and the method comprises the following steps: constructing a multi-layer parallel scheduling framework based on a heterogeneous supercomputing platform, and designing a molecular docking process of master-slave core asynchronous parallel computing, the invention provides a data access optimization scheme of a molecular docking key algorithm. According to the method, data reading, task scheduling and parallel computing in the molecular docking process are optimized from the aspect of nuclear memory access, the super-large-scale parallel drug virtual screening process is achieved, the computing speed is increased, and meanwhile the drug screening precision is improved. The I / O pressure of the system is reduced, the advantages of the heterogeneous many-core architecture are fully played, and the overall performance of the virtual drug screening system is improved.
Owner:OCEAN UNIV OF CHINA
Target screening method for coronavirus pneumonia resistance of sophora flower bud tea
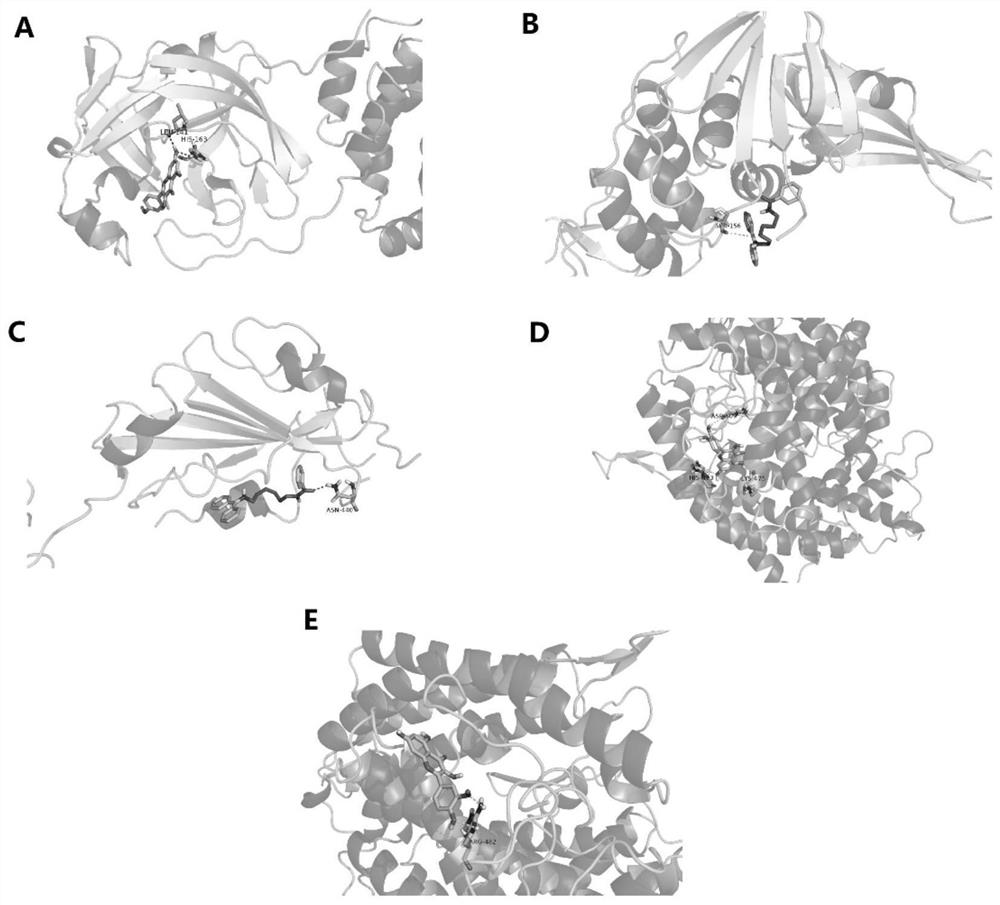
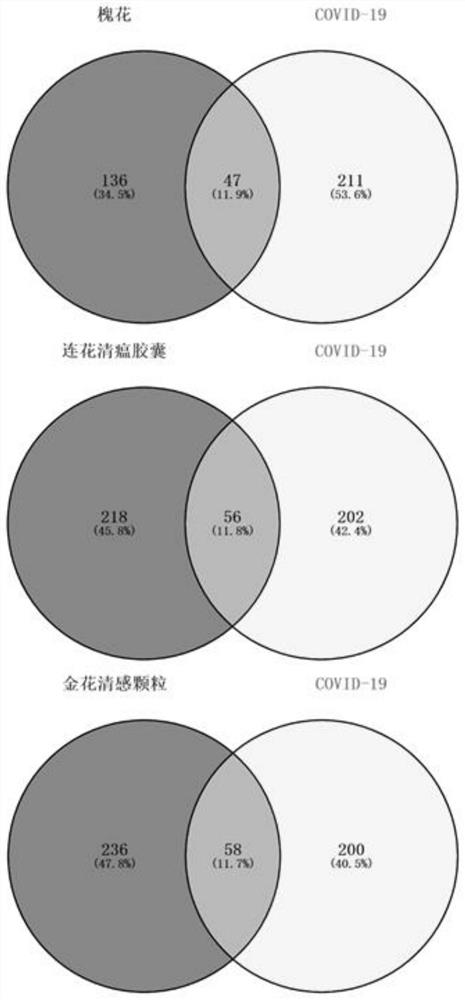
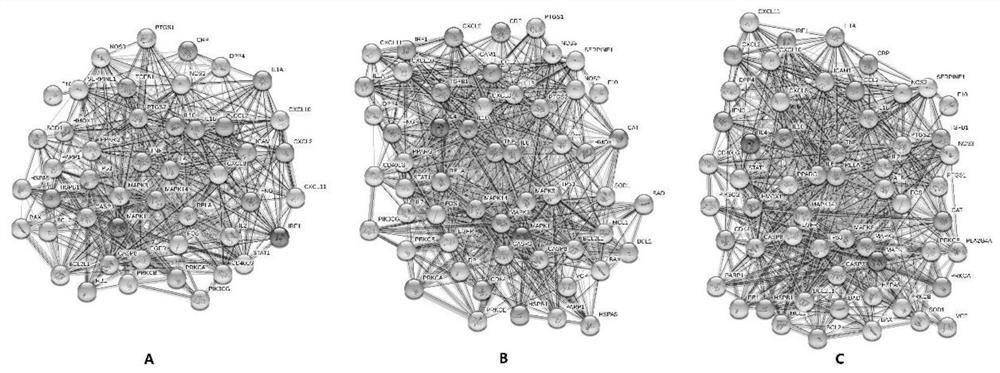
PendingCN112185479AHigh binding activityHigh affinityChemical property predictionMolecular entity identificationBiotechnologyDisease
The invention discloses a target screening method for coronavirus pneumonia resistance of sophora flower bud tea, and belongs to the technical field of medicines. The method comprises the following steps: screening active components and predicting a target; preparing a molecular docking ligand and a receptor; carrying out component target molecular docking to screen active components; predicting adisease target; constructing a compound-target-disease network; and carrying out key target function enrichment analysis. According to the research, a network pharmacology method and a molecular docking technology are adopted to explore active ingredients and potential targets of sophora flower buds for COVID-19, molecular docking verification is carried out, a component-target network is constructed, and the sophora flower bud tea is compared with lianhua Qingwen capsules and golden flower Qinggan particles to explore a mechanism action channel, and an idea is provided for subsequent basic experiments.
Owner:张升校
Application of Bevantol as AIBP inhibitor based on virtual screening
The invention discloses an application of Bevantol screened based on molecular docking and molecular dynamics simulation as an AIBP inhibitor, and the Bevantol screened based on simulation is used forinhibiting the interaction between AIBP and apoA-I, so cholesterol is inhibited from flowing out of cells. The invention also provides a simulation screening method for Bevantol screened based on molecular docking and molecular dynamics simulation as the AIBP inhibitor. The method comprises the following steps: obtaining an AIBP protein structure through homologous simulation, downloading a smallmolecular structure data set for docking, and performing a molecular docking process and molecular dynamics simulation in a ZINC database. According to the invention, the AIBP structure is obtained through homologous simulation, the virtual screening of FDA authenticated drugs is performed through molecular docking and molecular dynamics simulation on the basis of the new thought of old drugs, and it is found that Bevantol can be stably bonded to a bonding interface of AIBP and apoA-I. Or, cholesterol can be inhibited from flowing out of cells, and a new application of Bevantol is excavated.
Owner:NANTONG UNIVERSITY
Construction method of heparin C5 isomerase high-catalytic-activity strain
ActiveCN111304186AHigh catalytic activityBacteriaMicroorganism based processesEscherichia coliArginine
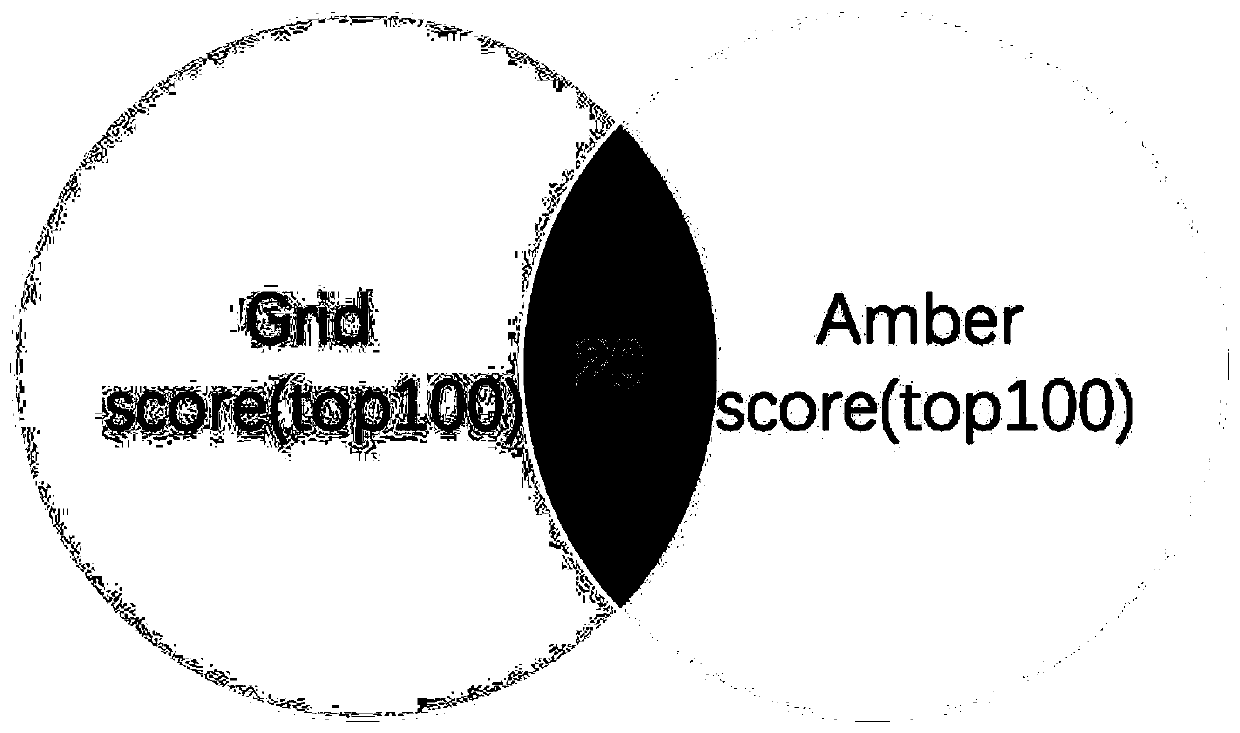
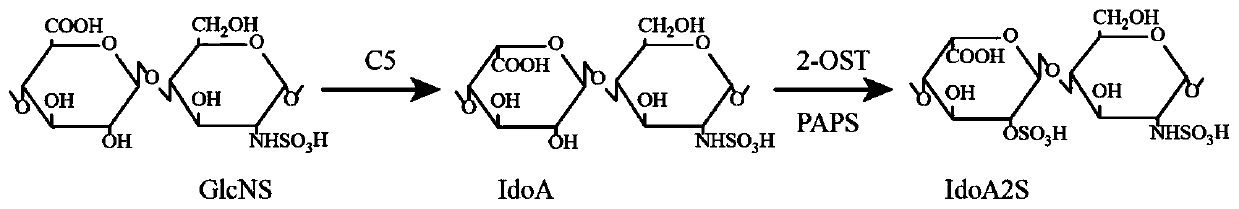
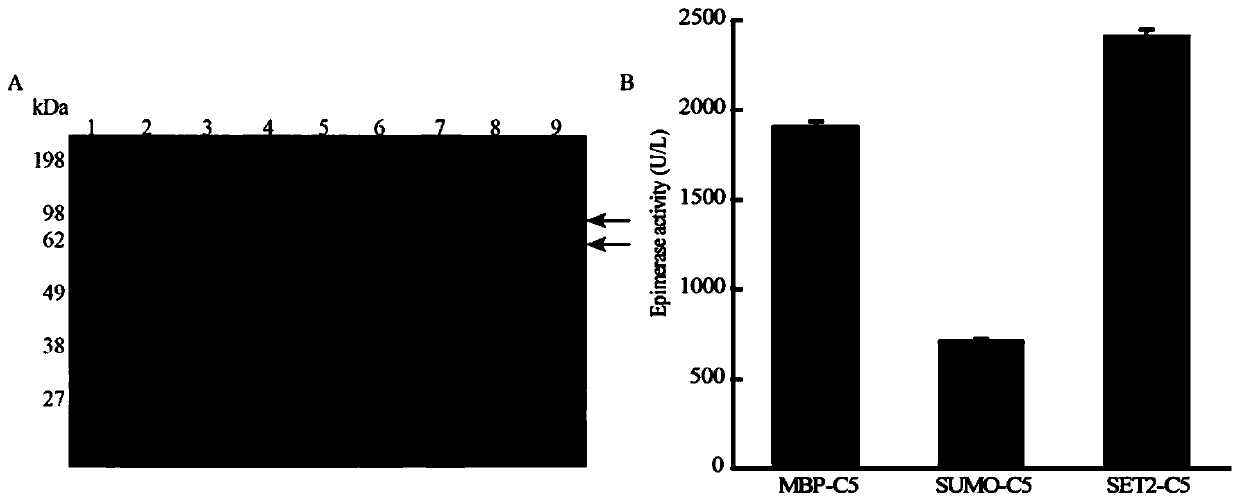
The invention discloses a construction method of a heparin C5 isomerase high-catalytic-activity strain, and belongs to the technical field of bioengineering. The method comprises the following steps:firstly, selecting Escherichia coli as host bacteria, then carrying out expression optimization on C5 isomerase by utilizing a way of fusing a solubilizing label at an N end, and further carrying outprotein engineering modification on an optimized C5 protein sequence through molecular docking so as to obtain a high-activity production strain. According to the invention, microbial cells are used for expressing the C5 obtained after mutation of 106 amino acids from valine to arginine for the first time, the catalytic activity of the obtained enzyme is 5.81 U / mL and is improved by 141% comparedwith that before mutation, and the specific enzyme activity is 145.14 U / mg and is improved by 128% compared with that before mutation.
Owner:JIANGNAN UNIV
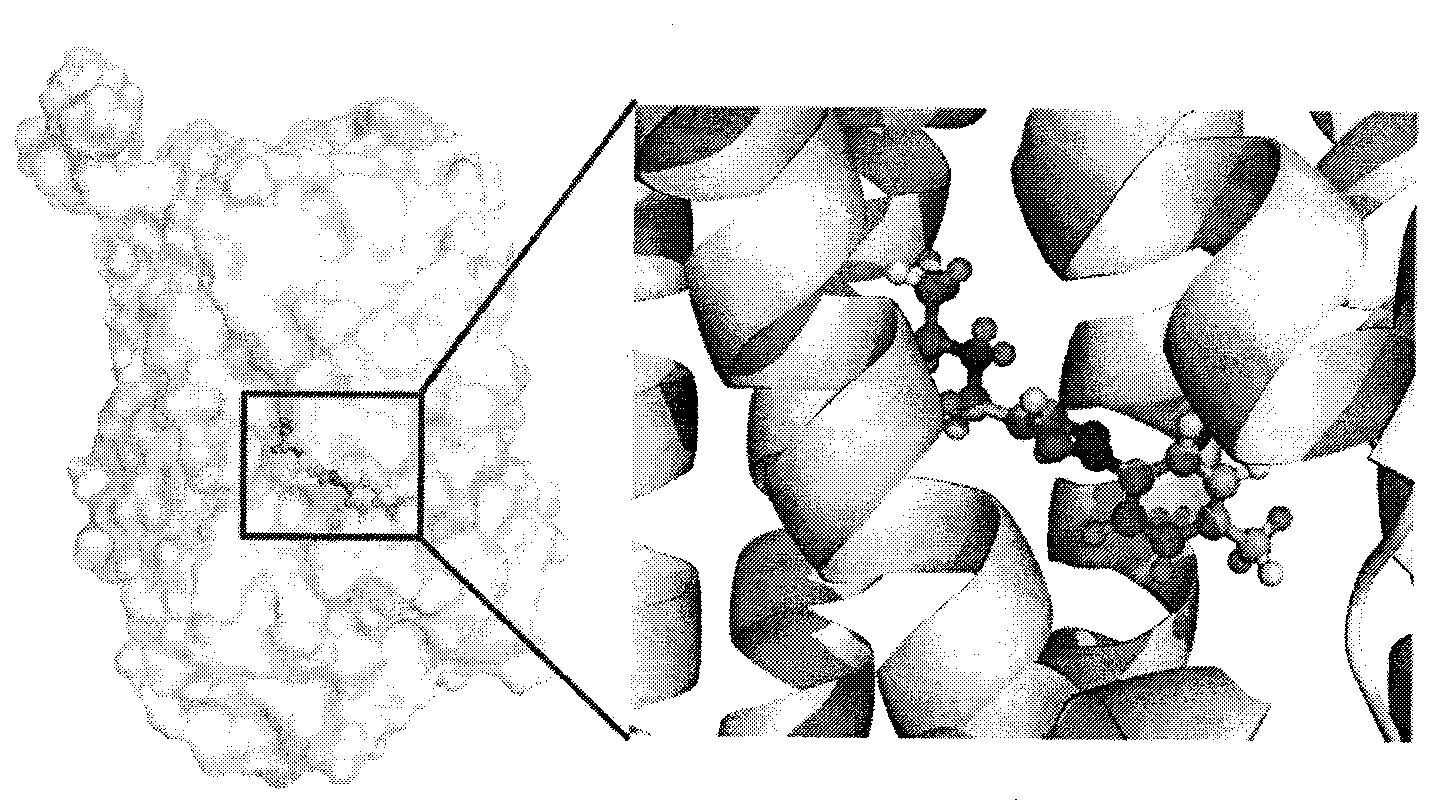
Target gene Rv0233 screened by antituberculous inhibitor and application
InactiveCN103123672AImprove hit rateReduce workloadMicrobiological testing/measurementMicroorganism based processesNucleotideBinding site
The invention belongs to the technical field of drug molecules and particularly relates to an application of mycobacterium tuberculosis gene Rv0233 in screening an antituberculous inhibitor target. The gene is characterized in that a nucleotide sequence of the gene is as shown in a sequence table: sequencer (SEQ) ID NO: 1, and a coded protein sequence of the gene is as shown in the sequence table: sequencer (SEQ) ID NO: 2. Eighteen small molecule compounds which have a high affinity with a binding site of natural substrate of Rv0233 screened out from a small molecular compound database through a molecular docking program aiming at an active centre of the natural substrate of the Rv0233 by the way of virtual screening. One bacteriostatic compound (pyrazole compound 51#) is obtained. The name of the compound is 2-({[1-(4-methyl pheny)-H-pyrazol-4-y1] methyl} amino) ethanol. The target gene evaluates the bacteriostatic effect of the compound on the mycobacterium tuberculosis.
Owner:HUAZHONG AGRI UNIV
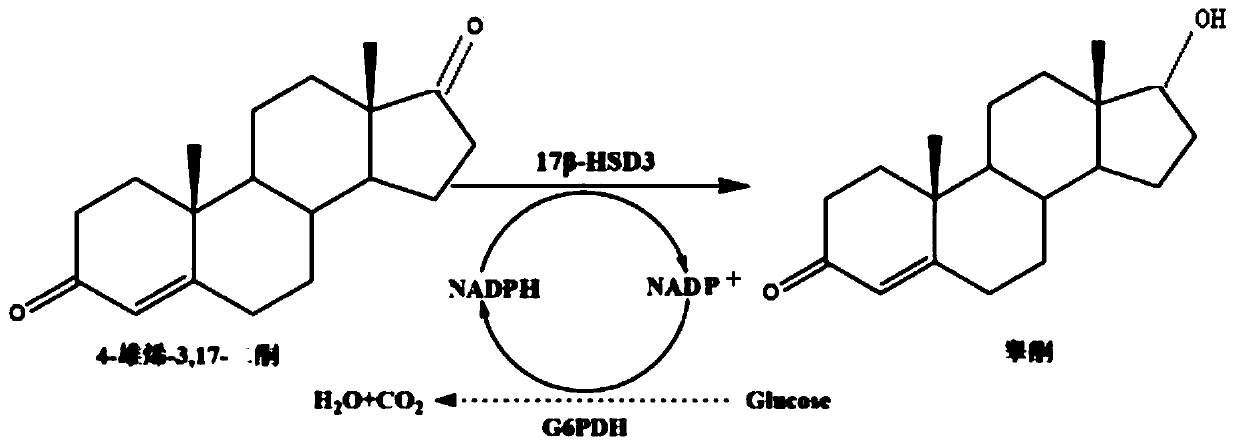
17 beta-hydroxysteroid hydroxylase 3 mutant enzyme, coding gene and engineering bacteria
ActiveCN111454919AHigh catalytic activityIncrease enzyme activityBacteriaMicroorganism based processesHormone drugHomosteroids
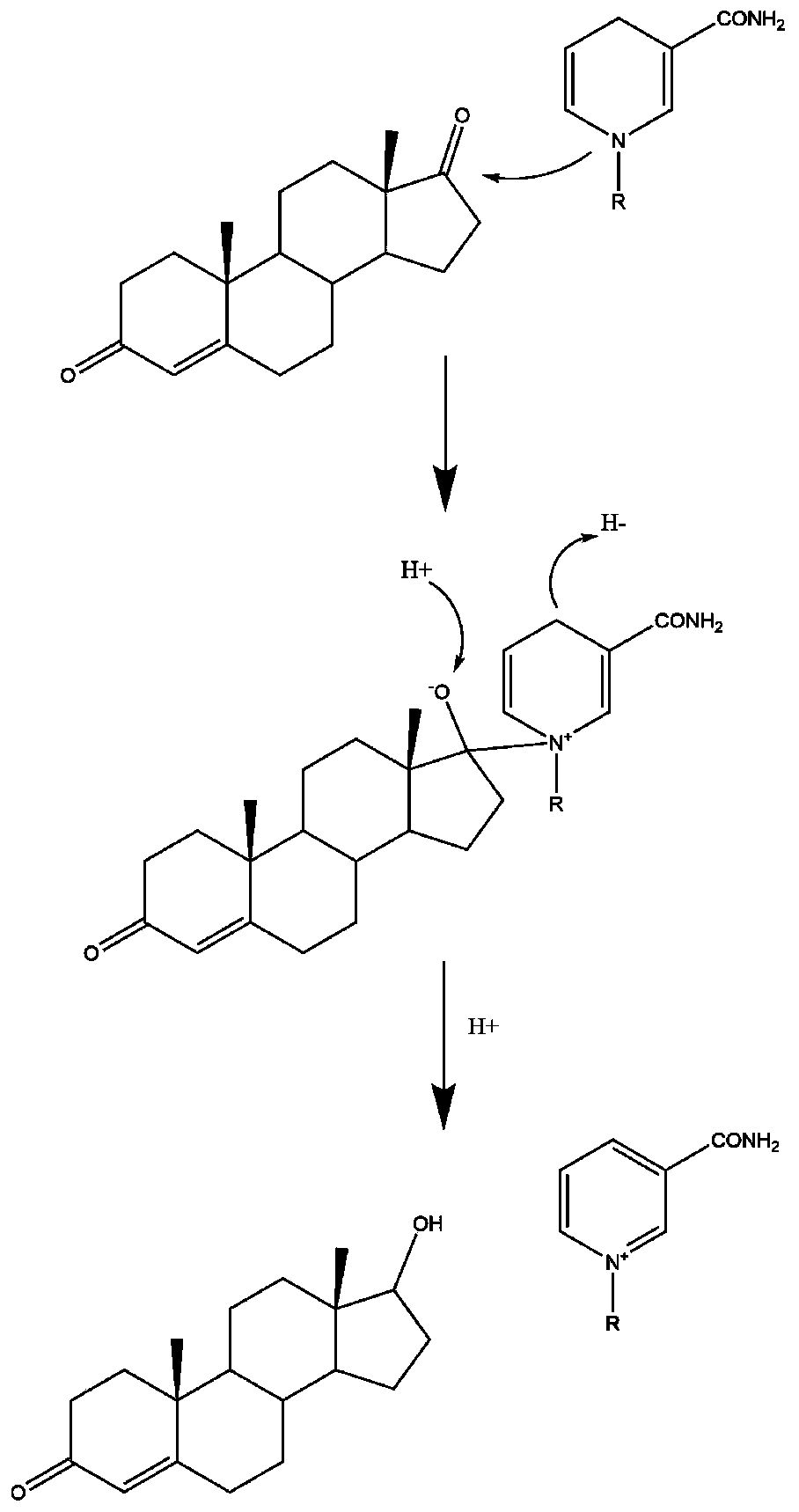
The invention discloses a 17 beta-hydroxysteroid hydroxylase 3 mutant enzyme, a coding gene and engineering bacteria. The 17 beta-hydroxysteroid hydroxylase 3 mutant enzyme is abbreviated as 17 beta-HSD3<G186R / Y195W>, and an amino acid sequence of the 17 beta-hydroxysteroid hydroxylase 3 mutant enzyme is as shown in SEQ ID NO.1. 17 beta-hydroxysteroid hydroxylase 3 is subjected to molecular modification by adopting a rational design technology, the 17 beta-hydroxysteroid hydroxylase 3 mutant enzyme with improved affinity with a substrate is obtained through methods of homologous modeling, molecular docking, binding energy calculation and the like, the enzyme activity is improved by 1.66 times as compared with the original enzyme activity, and by utilizing the strategy, the catalytic activity of the 17 beta-hydroxysteroid hydroxylase 3 can be obviously improved. The 17 beta-hydroxysteroid hydroxylase 3 mutant enzyme is coded, so that the yield of testosterone is up to 3.95 g / L, and technical support and data reference are provided for large-scale biological preparation of the testosterone which is a male hormone drug.
Owner:TIANJIN UNIV
Virtual drug screening method and device based on molecular docking
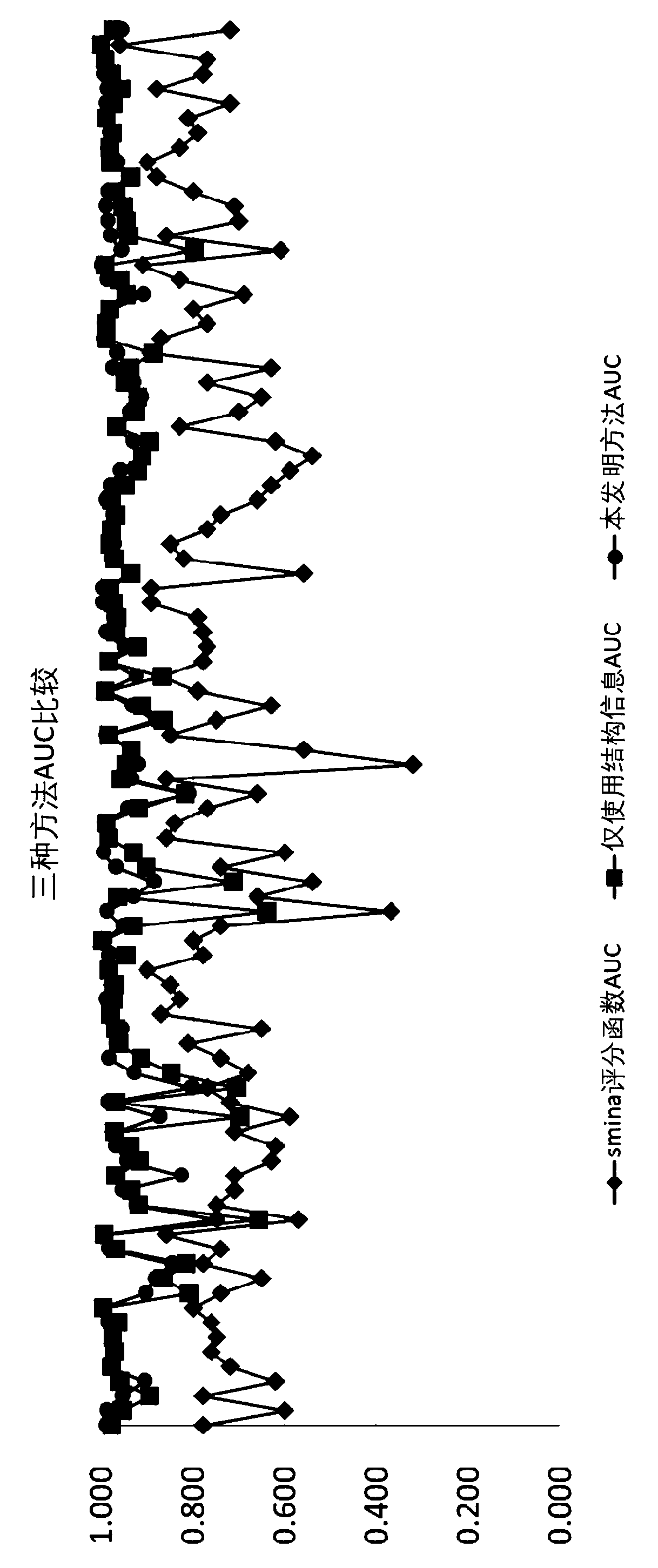
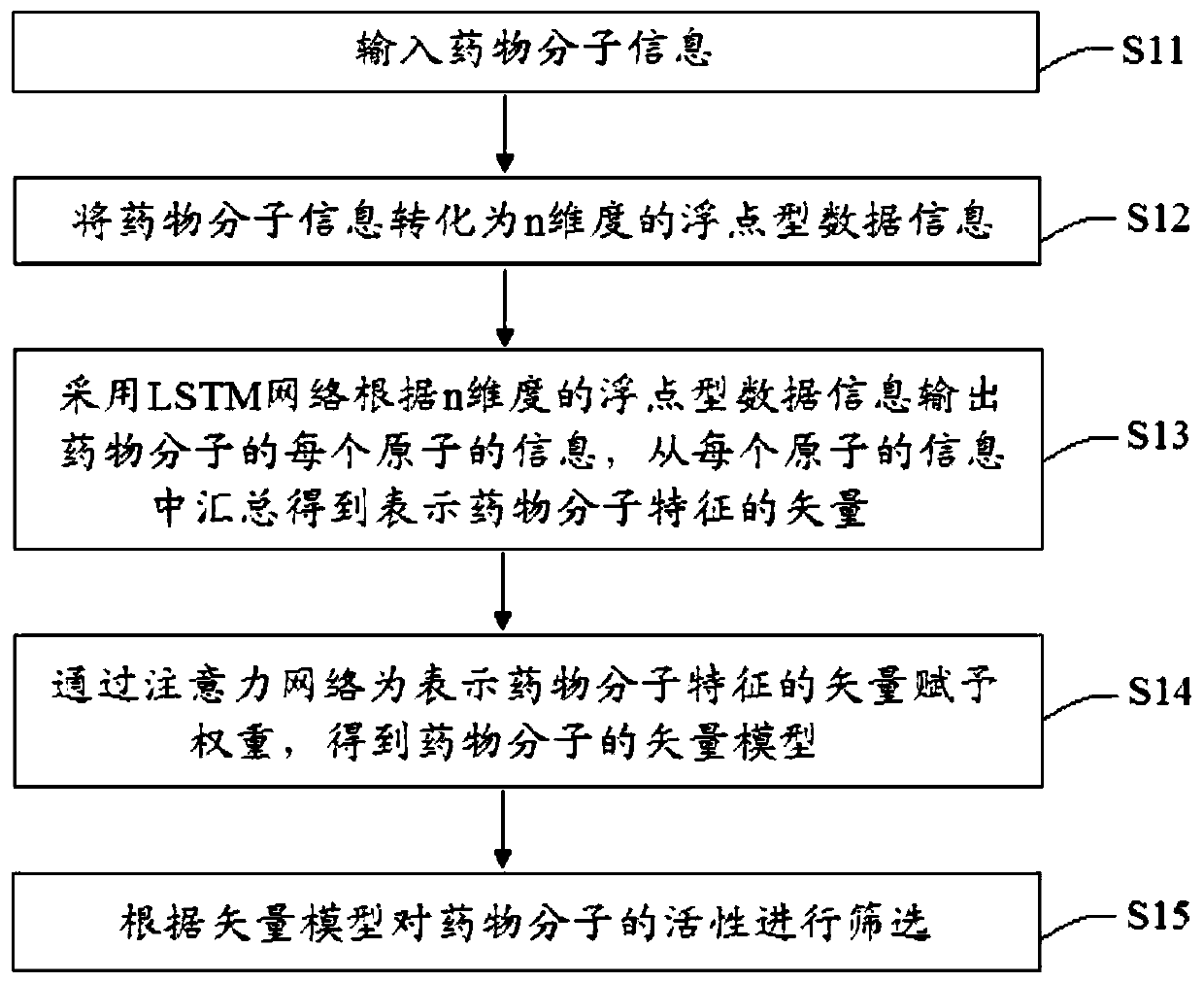
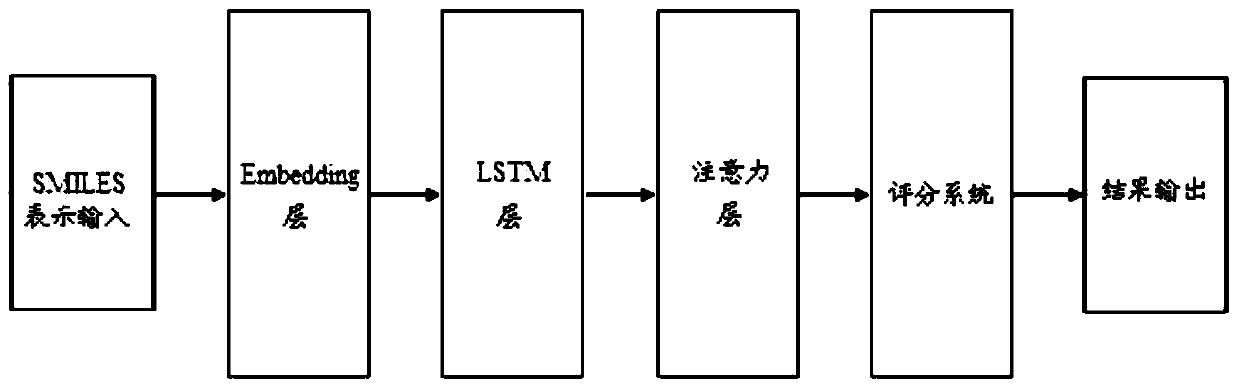
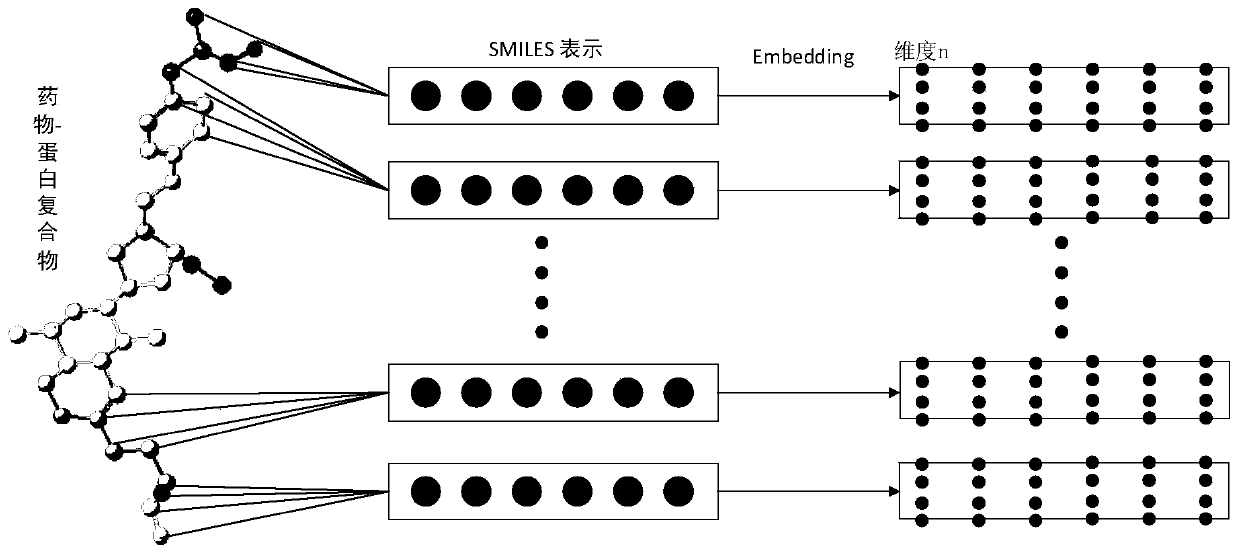
PendingCN111199779AAlgorithm AUC value improvementImprove robustnessMolecular designNeural architecturesPharmaceutical drugBiochemistry
The invention provides a virtual drug screening method and device based on molecular docking. The method comprises the following steps: inputting drug molecular information; converting the drug molecule information into n-dimensional floating point type data information; outputting the information of each atom of the drug molecule according to the n-dimensional floating point type data informationby adopting an LSTM network, and summarizing the information of each atom to obtain a vector representing the characteristics of the drug molecule; assigning a weight to the vector representing the characteristics of the drug molecule through an attention network to obtain a vector model of the drug molecule; and screening the activity of the drug molecules according to the vector model. According to the scheme, the AUC value can be increased by 2%, and higher robustness is achieved.
Owner:中科曙光国际信息产业有限公司
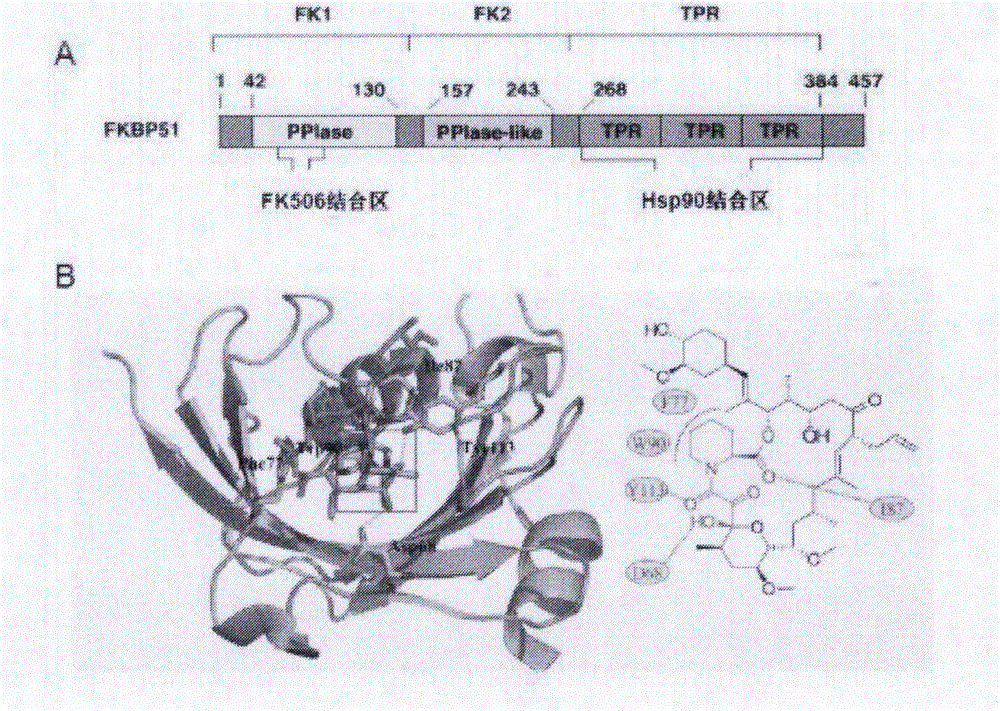
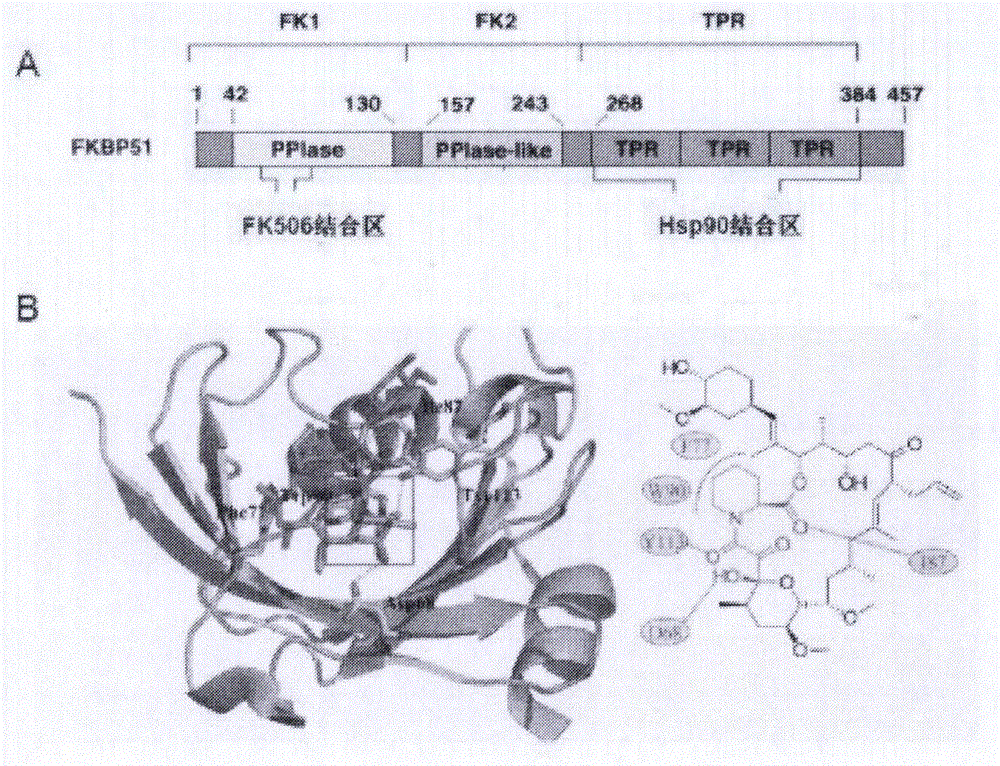
Lead compound for targeted human FKBP51 protein and screening method and application thereof
InactiveCN105902537AStrong off-target effectCompound screeningOrganic active ingredientsProstate cancer cellProtein target
The invention provides a lead compound for a targeted human FKBP51 protein and a screening method and application thereof, belonging to the technical field of biochemical pharmacy. According to the lead compound and the screening method and application thereof, an FK1 structural domain of FKBP51 is used as a receptor; FK506 of the FKBP51 is selected to be combined with a sack; molecular docking is performed by using a Glide program; 150 small molecular compounds are obtained by virtual screening from more than 1.5 million of compounds and are used for fluorescence quenching experiments; according to the experiments, a combination action of the compounds and the FK1 is verified, and the binding property of a target protein and compounds with a strong docking effect is researched; two kinds of cells LNCaP and DU145 are selected to verify the cell viability of screened small molecular compounds resisting prostate cancer; meanwhile, the structural biology research on a compound formed by the FKBP51 and small molecular lead compounds is performed. According to the lead compound provided by the invention, a cell model and a mouse model of CRPC (Castration Resistant Prostate Cancer) are established, the effectiveness of the lead compound to the CRPC is evaluated, and an action mechanism and a rule of the lead compound and the target protein are explained at the molecular level; a foundation is provided for researching and developing new drugs for treating common prostate cancer and the CRPC.
Owner:LANZHOU UNIVERSITY
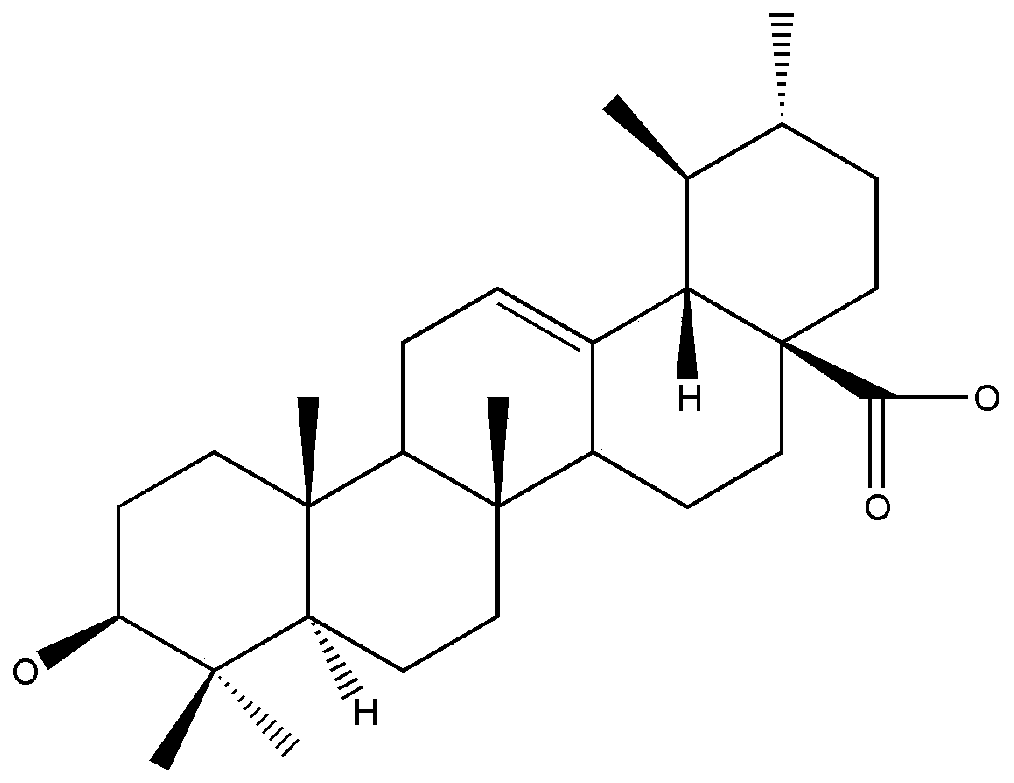
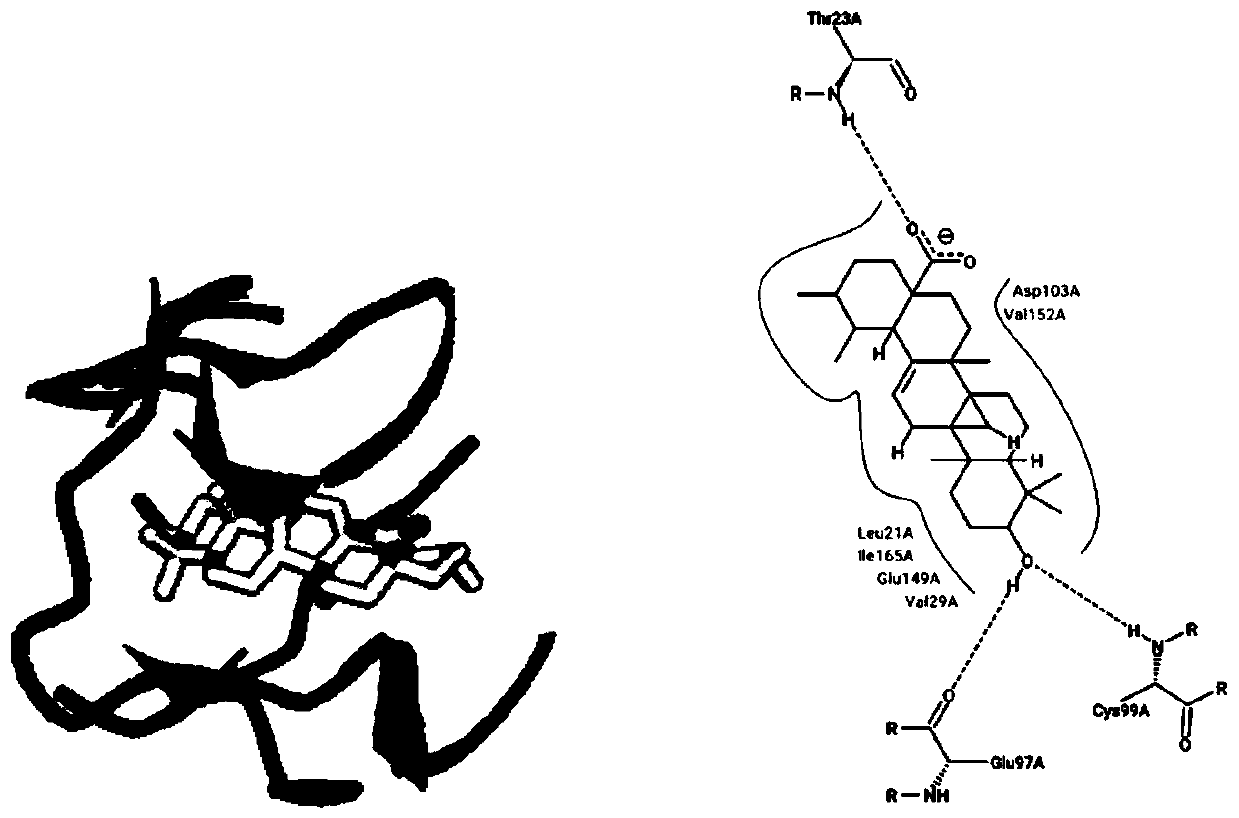
Design and preparation method of epoxide hydrolase mutant
ActiveCN104531630AHigh enantioselectivityHigh catalytic efficiencyHydrolasesFermentationBiotechnologyEpoxide metabolism
The invention provides design and construction of AuEH2G218S and AuEH2S247Y mutant enzymes with high enantio-selectivity. The mutation sites are finally determined by sequence homology analysis, homologous modeling, molecular docking and dynamic simulation. Mutant genes obtained by a megaprimer RPC technology are named Aueh2G218S and Aueh2S247Y. The invention further discloses a method for construction and efficient expression of mutant engineering bacteria; and the prepared mutant enzymes have the characteristics of high enantio-selectivity and catalytic activity, and have relatively large industrial production potential.
Owner:JIANGNAN UNIV
Virtual screening method of targeted IKK beta drug
The embodiment of the invention discloses a virtual screening method of a targeted IKK beta drug, and belongs to the technical field of medicinal chemistry. The method comprises the following steps: by taking ursolic acid as a ligand and IKKbeta (PDB ID: 4KIK) as receptor protein, analyzing a binding site and binding energy of ursolic acid and IKK beta and a binding side chain group of ursolic acid by adopting molecular docking software; analyzing a binding site and binding energy of the modified ursolic acid and IKK beta by adopting molecular docking software; preliminarily screening out a side chain group with a relatively good binding effect; and carrying out molecular modification on ursolic acid to obtain an ursolic acid derivative, and adopting molecular docking software to preliminarily determine a targeted IKKbeta drug. Based on the principle and method of molecular docking, ursolic acid is subjected to side chain group modification, the ursolic acid derivative with good docking capacity is screened out, and the ursolic acid derivative has good application value and prospect in the field of development of antitumor drugs with ursolic acid as a primer.
Owner:SUZHOU UNIV OF SCI & TECH
Regioselective bacterium nitroreductase gene and application thereof
InactiveCN104099351AHigh biological toxicityHigh reducing activityOxidoreductasesFermentationDiseaseEscherichia coli
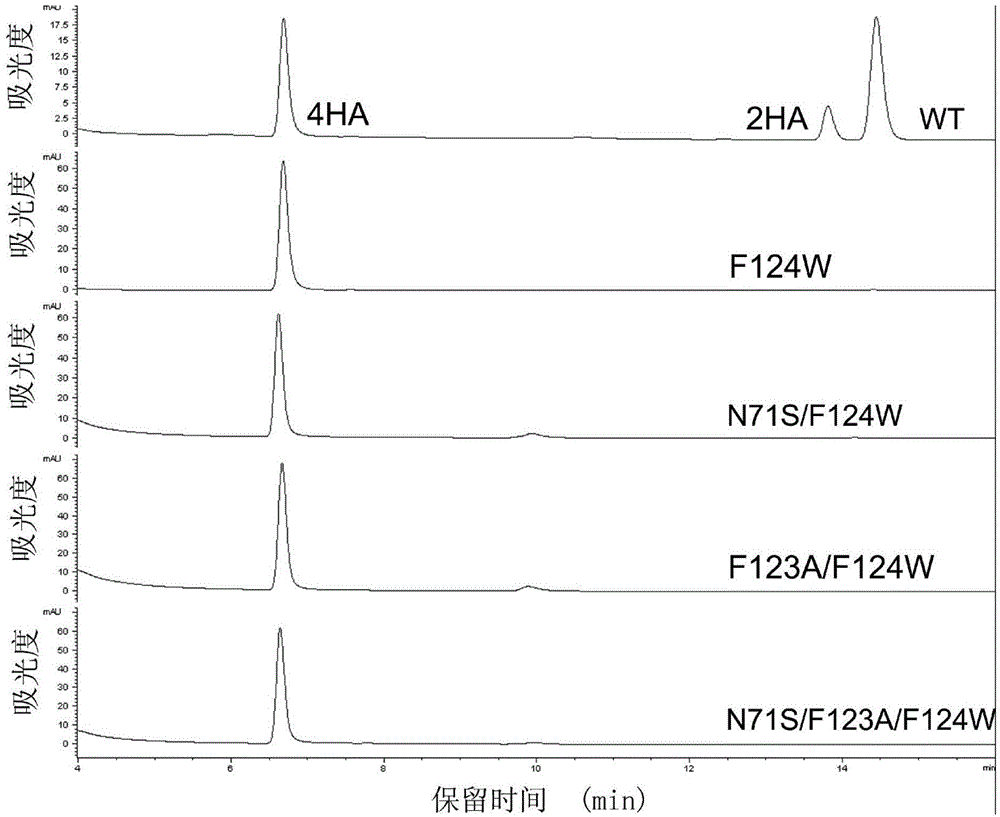
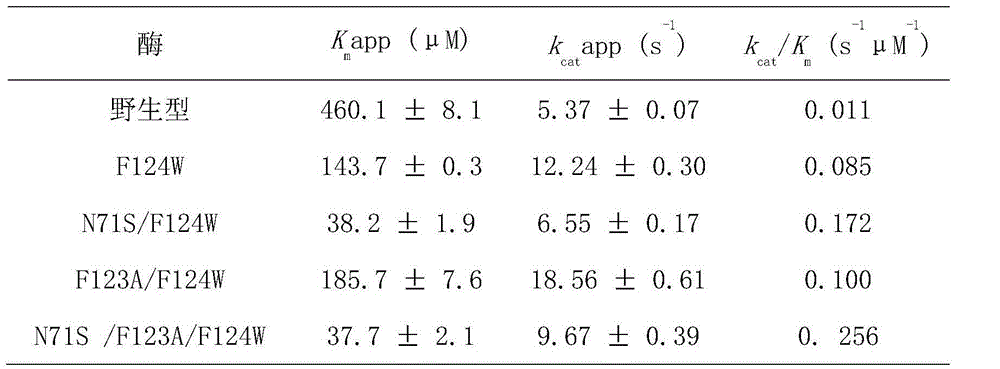
The invention discloses a mutant bacterium nitroreductase gene and an application thereof and belongs to the field of enzyme engineering. On the basis of protein three-dimensional structures and molecular docking, phenylalanine in the 124 position of Escherichia coli nitroreductase NfsB interacts with side chain groups of a multi-nitro substrate, so that positioning of the substrate in an activity pocket is affected. Mutants F124W, N71S / F124W, F123A / F124W and N71S / F123A / F124W can catalyze the specificity of a prodrug CB1954 for treating cancers to produce a 4-NHOH product with high biotoxicity. Meanwhile, the catalytic activity of the mutants is improved by 7-24 times when being compared with that of wild type enzymes, and the N71S / F123A / F124W is the most significant. The nitroreductase mutants with specific reduction selectivity have the good application prospect in aspects of treatment of cancers and other diseases.
Owner:DALIAN UNIV OF TECH
Molecular docking result screening method based on positive compound residue contribution similarity
ActiveCN113380320AImprove screening rateHigh precisionMolecular designComputational theoretical chemistryChemical compoundPharmaceutical drug
The invention relates to a molecular docking result screening method based on positive compound residue contribution similarity, and belongs to the technical field of drug screening, and the method comprises the steps of constructing a positive compound library, optimizing target spots and compound structures, and optimizing a docking result screening method. According to the method disclosed by the invention, the screening probability of active compounds is increased in the process of finding potential patent medicine compounds, so that the precision of the screening method is fundamentally improved, the accuracy of screening the potential patent medicine compounds is improved, and the calculation cost and the time cost are greatly saved.
Owner:OCEAN UNIV OF CHINA
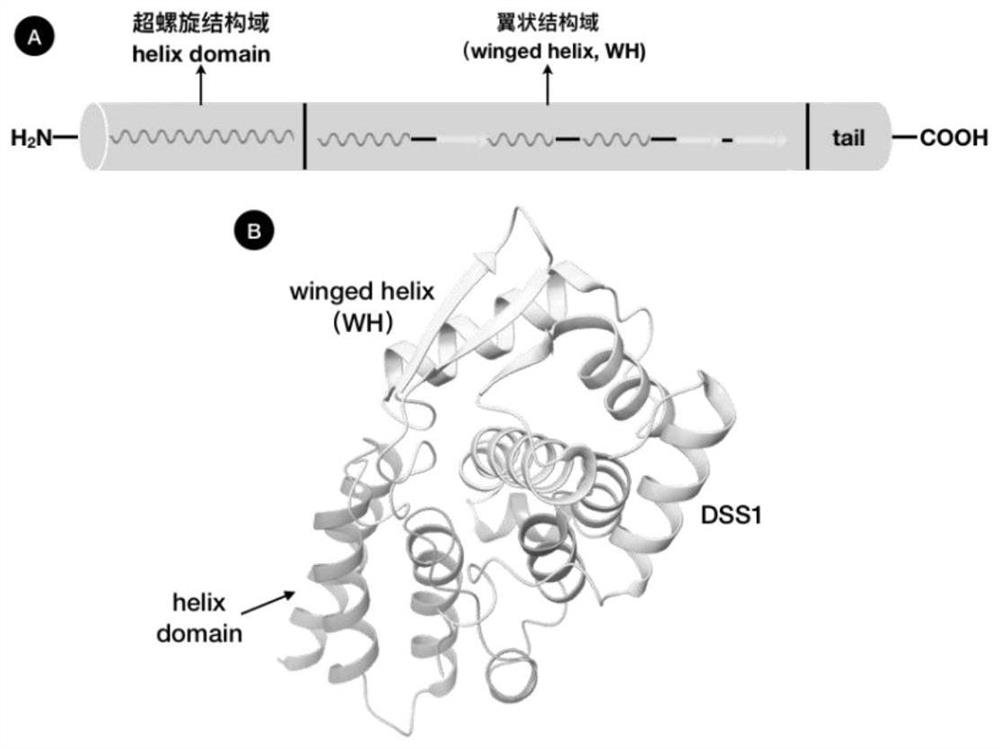
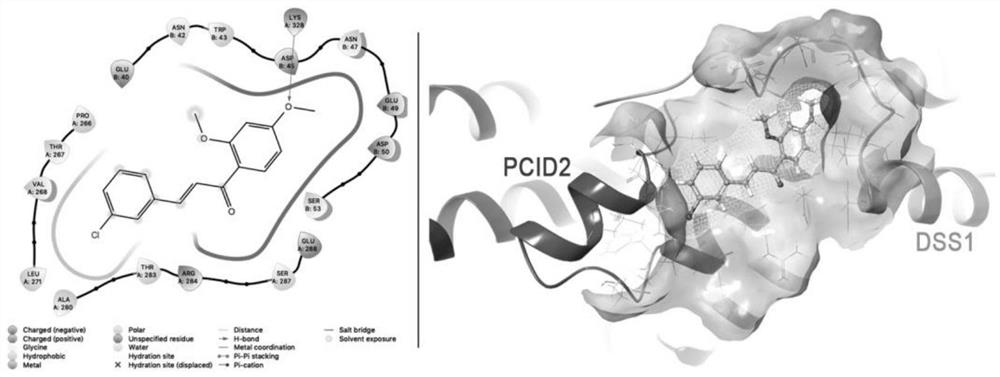
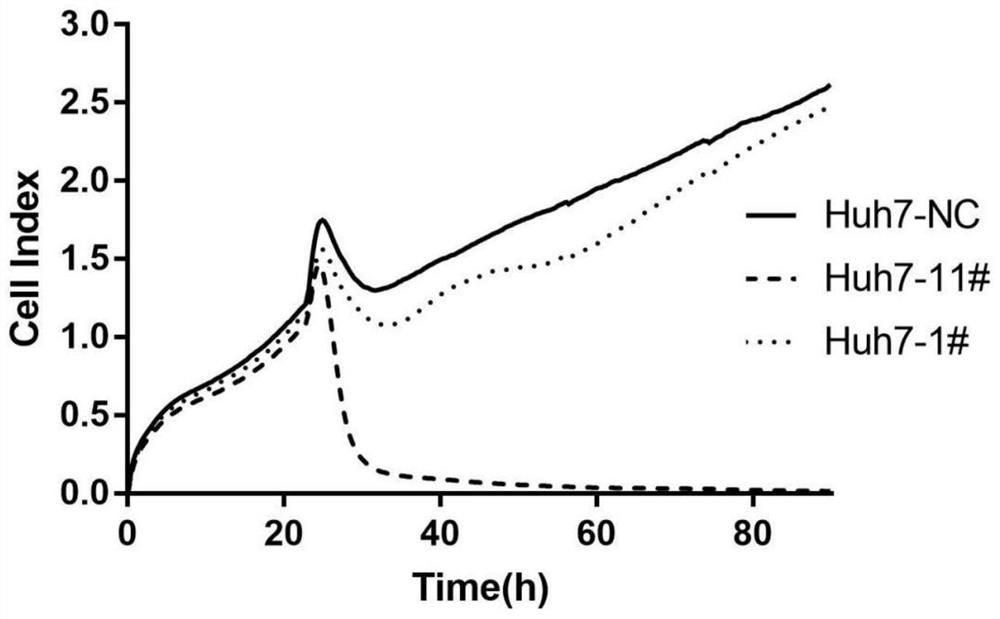
Application of human PCID2 protein in preparation or screening of antitumor drugs and compound with antitumor activity
PendingCN114426470APrevent proliferationInhibitory activityOrganic chemistryAntineoplastic agentsHumaninProtein target
The invention belongs to the technical field of biotechnology and gene therapy, and particularly relates to application of human PCID2 protein in preparation or screening of anti-tumor drugs and an anti-tumor compound. It is found that the PCID2 gene promotes tumor cell proliferation and regulates the cell cycle, is a key molecule for regulating tumor generation and development, and can be used as a target protein for preparing or screening anti-tumor drugs; meanwhile, human PCID2 protein is used as target protein, a class of PCID2 protein targeting antitumor compounds are screened out through a molecular docking technology, and the compounds can significantly inhibit tumor cell proliferation and promote tumor cell apoptosis.
Owner:THE FIRST HOSPITAL OF LANZHOU UNIV
Application of Irinotecan based on simulation screening as AIBP inhibitor
The invention provides an application of Irinotecan based on molecular docking and molecular dynamics simulation screening as an AIBP inhibitor, and the Irinotecan is used for inhibiting interaction between AIBP and apoA-I, so that cholesterol is inhibited from flowing out of cells. The invention also provides a method for Irinotecan as an AIBP inhibitor based on molecular docking and molecular dynamics simulation screening. The method comprises the following steps: obtaining an AIBP protein structure through homologous simulation, and downloading a small molecular structure data set for docking, a molecular docking process and molecular dynamics simulation in a ZINC database. According to the method, the structure of AIBP is obtained through homologous simulation, the virtual screening isconducted on FDA authenticated drugs through molecular docking and molecular dynamics simulation on the basis of the new thought of old drugs, and it is found that Irinotecan can be stably bonded toa bonding interface of AIBP and apoA-I; or cholesterol can be inhibited from flowing out of cells, and the new application of the Irinotecan is excavated.
Owner:NANTONG UNIVERSITY
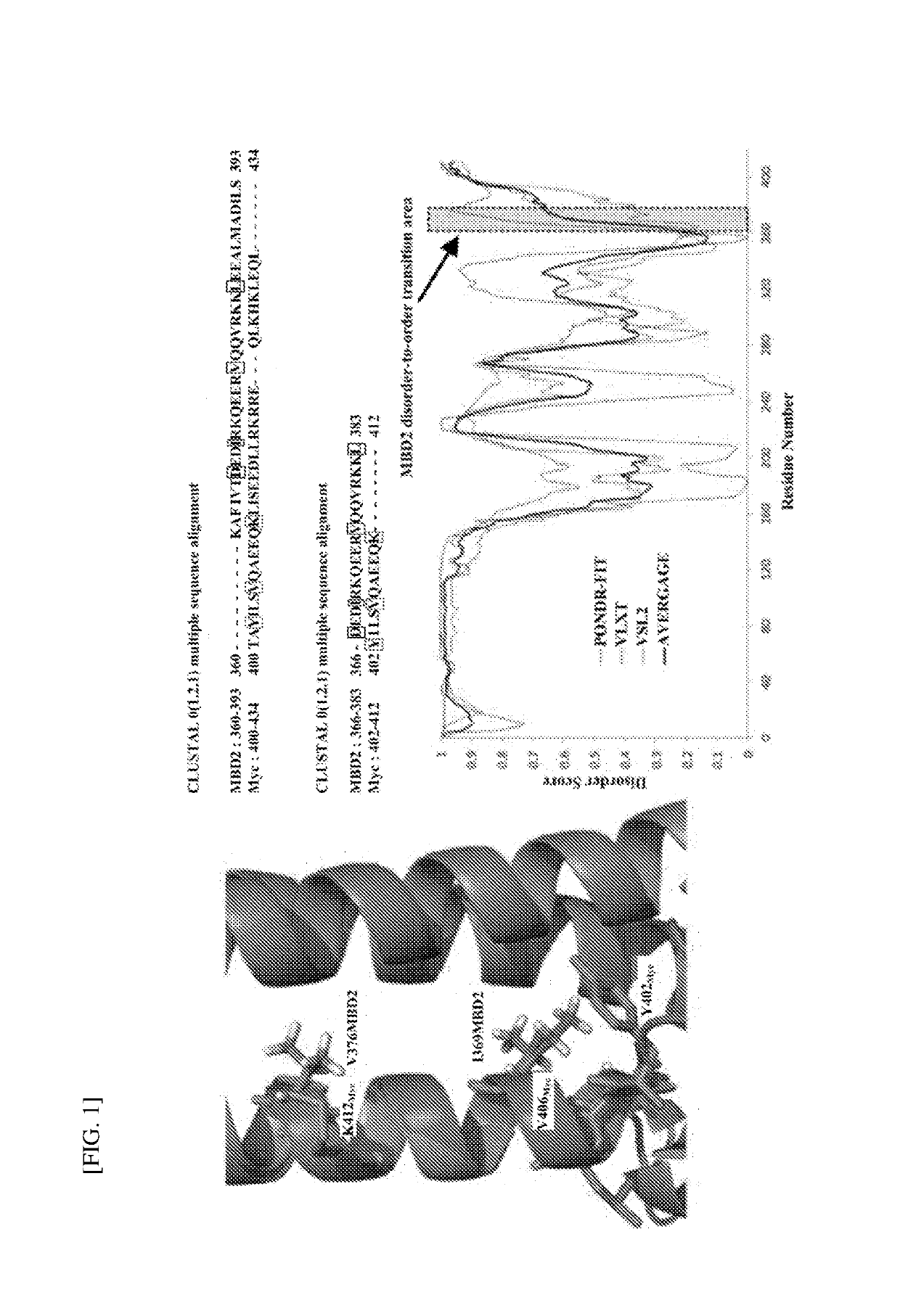
Method of discovering new drug candidate targeting disorder-to-order transition region and apparatus for discovering new drug candidate
PendingUS20190279737A1Low costShorten the timeCompound screeningApoptosis detectionProtein targetChemical compound
A method of discovering a drug candidate, comprising: a step in which a computer device uses bioinformatics to determine a disorder-to-order transition region of a target protein; a step in which the computer device performs molecular docking on the disorder-to-order transition region in conjunction with a library of specific compounds to select first candidate compounds capable of binding to the disorder-to-order transition region from among the compound library; and a step in which the computer device performs a molecular dynamics simulation for the first candidate compounds and the disorder-to-order transition region to select a second candidate compound from among the first candidate compounds.
Owner:IUCF HYU (IND UNIV COOP FOUNDATION HANYANG UNIV)
Lead compound using human FKBP51 protein as target as well as screening method and application thereof
ActiveCN105963301AStrong off-target effectOrganic active ingredientsOrganic chemistryLead compoundMacromolecular docking
The invention provides a lead compound using human FKBP51 protein as a target as well as a screening method and an application thereof, and belongs to the field of biochemical pharmacy technology. A FK1 domain of the FKBP51 protein is used as a receptor, a FK506 binding pocket of the receptor is selected, a Glide program is used for carrying out molecular docking, 150 small molecule compounds are obtained by virtual screening from more than 1.50 million compounds, and fluorescence quenching experiments are carried out for checking binding effects between the compounds and FK1; binding characteristics between the target protein and the compounds with high docking effects are researched, activities of the screened small molecule compounds against prostate cancer cells are checked by using two kinds of selected cells including LNCaP and DU145, and at the same time structural biology researches are carried out for compounds formed by FKBP51 and the small molecule lead compounds. The invention also establishes a cell model and a mice model of CRPC, effectiveness of the lead compounds on CRPC are evaluated, mechanisms and rules of action between the lead compounds and the target protein on molecular level are illustrated, and a foundation is provided for research and development of new drugs for treating common prostate cancer and CRPC.
Owner:LANZHOU UNIVERSITY
Docked aptamer eab biosensors
InactiveCN110998326AReduce dependenceMaterial analysis by electric/magnetic meansGenetic engineeringAptamerAnalyte
The invention describes electrochemical aptamer-based biosensing devices and methods that are configured to produce a detectible signal upon target analyte interaction, with reduced reliance on a conformational change by the aptamer. The disclosure includes docked aptamer EAB sensors for measuring the presence of a target analyte in a biofluid sample. The sensors include an electrode capable of sensing redox events, and aptamer sensing elements with aptamers selected to interact with a target analyte. Each aptamer sensing element includes a molecular docking structure attached to the electrode, and an analyte capture complex that includes an aptamer releasably bound to the docking structure, and an electroactive redox moiety. Upon aptamer binding with a target analyte, the analyte capturecomplex separates from the docking structure. The separation of the analyte capture complex from the docking structure produces a positional change in the redox moiety that is detectable by the sensing device on interrogation of the electrode.
Owner:ECCRINE SYST
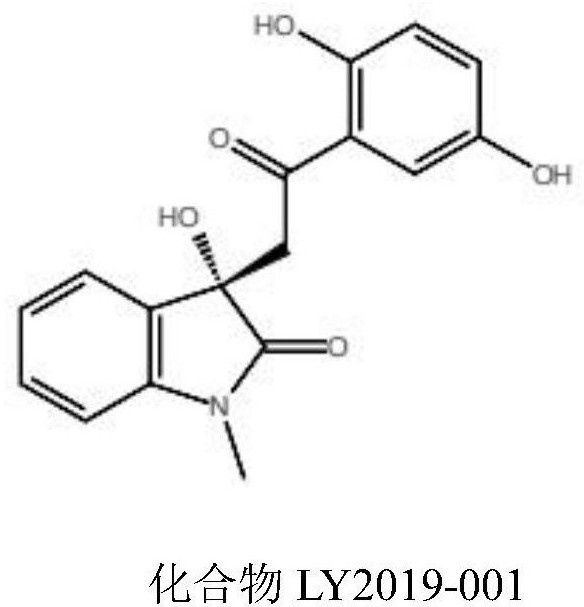
Humanized hexokinase 2 small-molecule inhibitor and application thereof
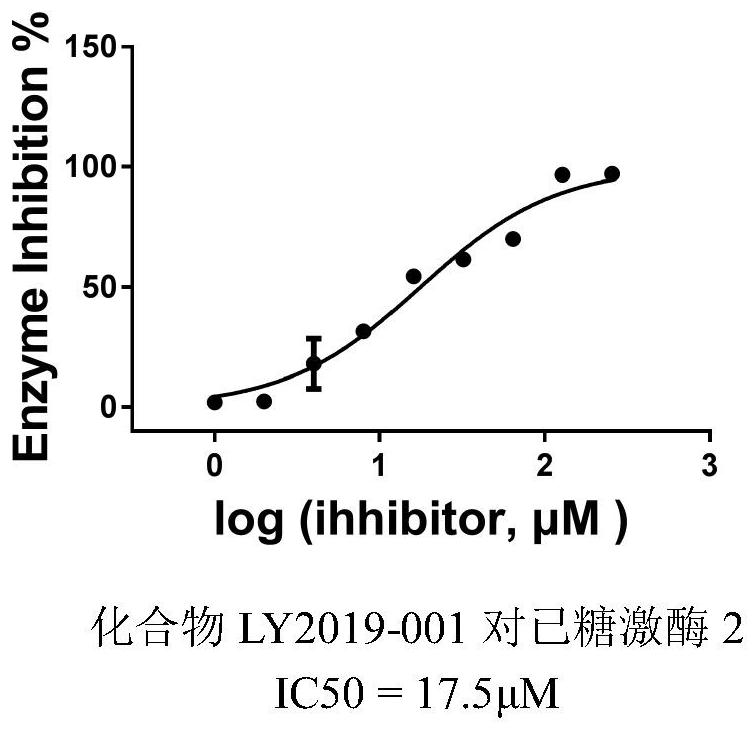
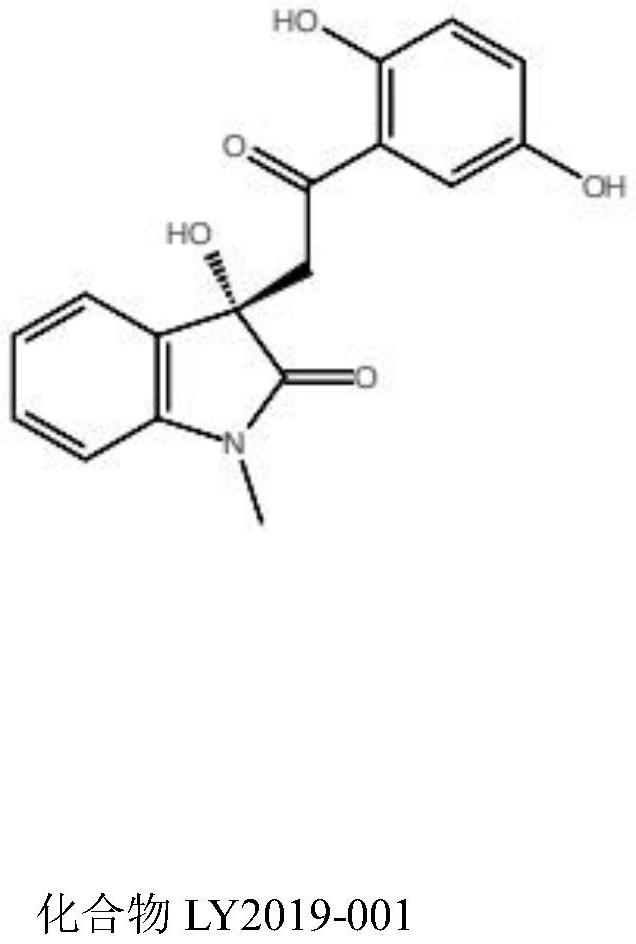
InactiveCN112891342AGrowth inhibitionOrganic active ingredientsAntineoplastic agentsDiseaseProstate cancer
The invention provides a small-molecule inhibitor taking human hexokinase 2 as a target spot and application of the small-molecule inhibitor, and belongs to the technical field of pharmacy. According to the invention, hexokinase 2 is used as a receptor, an inhibitor binding pocket of hexokinase 2 is selected, molecular docking is carried out by using a Glide program, virtual screening is carried out from more than 1,600,000 compounds to obtain the small-molecule inhibitor LY2019-001 of targeting human hexokinase 2, and enzyme activity inhibition experiments and MTT cell proliferation inhibition experiments are carried out; and the inhibition effect of the compound LY2019-001 on hexokinase 2 and the growth inhibition effect of the compound LY2019-001 on cervical cancer cells HeLa, liver cancer cells HepG2 and lung cancer cells A549 are verified. The found small molecules and pharmaceutical salts thereof can provide a basis for new drug research and development for treating diseases related to hexokinase 2, such as liver cancer, lung cancer, endometrial cancer, breast cancer, ovarian cancer, pancreatic cancer or prostate cancer. The structure of the lead compound can be further optimized, and the lead compound has a relatively good application prospect.
Owner:LANZHOU UNIVERSITY
Genetically engineered bacterium for efficiently synthesizing alpha-arbutin and construction method and application of genetically engineered bacterium
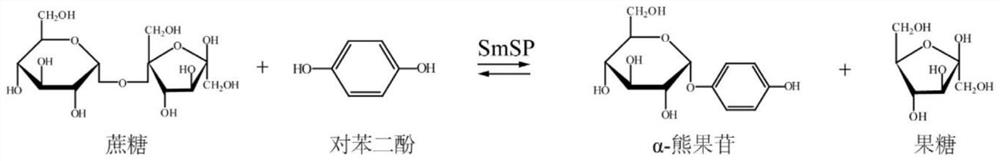
ActiveCN112375724AHigh protein expressionIncrease productionBacteriaMicroorganism based processesSucrose phosphorylasePhosphorylation
The invention discloses a genetically engineered bacterium for efficiently synthesizing alpha-arbutin and a construction method and application of the genetically engineered bacterium, and belongs tothe technical field of genetic engineering. Alpha-arbutin is efficiently produced by whole-cell catalysis of recombinant bacillus subtilis for expressing sucrose phosphorylase (SmSP), and a new thought and a new method are provided for production of alpha-arbutin. The protein mutant SmSPI336L with improved SmSP enzyme activity is screened through molecular docking and site-specific saturation mutagenesis, the protein expression quantity of SmSP is increased by increasing the copy number of SmSPI336L on a genome, and the yield of alpha-arbutin is greatly increased by optimizing the catalytic condition of a BS-3SmSPI336L strain. When the concentration of a substrate HQ is 50 g / L, the sucrose concentration is 310.9 g / L, the thallus concentration OD600 is 40, the yield of alpha-arbutin reaches115.8 g / L when a catalytic system catalyzes for 20 hours in a shake flask at 30 DEG C and 220 rpm, and the molar conversion rate of the substrate HQ is 93.6%.
Owner:JIANGNAN UNIV
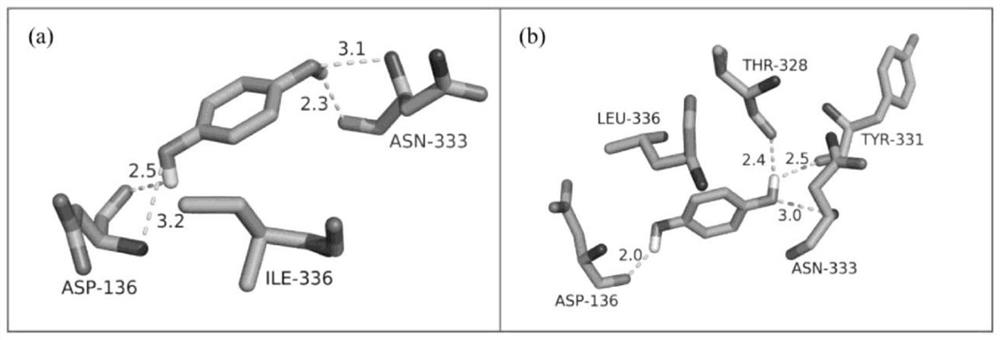
Application of plant essential oil and perfume compound in preparation of sanitary product for inhibiting infective ability of novel corona-virus
InactiveCN112741121AHigh potential activityImprove bindingBiocideDisinfectantsChemical compoundBiochemistry
The invention discloses application of a series of plant essential oil and perfume compounds in preparation of incense products for inhibiting infectivity of novel corona-virus. A computer molecular docking test shows that main compounds contained in the plant essential oil have strong binding force with a potential target S protein of new corona-virus, the binding energy [delta]G of about 210 compounds and the S protein is smaller than or equal to -5.0 kcal / mol, and steric hindrance is formed for binding of ACE2 after the S protein is bound with the compounds. Moreover, the total amount of components with binding energy [delta]G of compounds and S protein being less than or equal to -5.0 kcal / mol in the essential oil is more than 20%, so that the plant essential oil has relatively high potential activity in the aspect of inhibiting SARS-CoV-2 from infecting host cells or self-replication, can reduce or inhibit the infectivity of corona-virus, and is expected to be applied to blocking the corona-virus epidemic situation when being applied to the incense.
Owner:GUANGXI INST OF BOTANY THE CHINESE ACAD OF SCI
Application of tripterine in preparation of anti-coronavirus products
PendingCN113813264AThe mechanism of action is clearEnhanced inhibitory effectOrganic active ingredientsAntiviralsTripterineBiochemistry
The invention discloses application of tripterine in preparation of anti-coronavirus products, and belongs to application in the field of medicines. The inhibition effect of the tripterine on the S protein and ACE2 protein of novel coronavirus COVID-19 is proved by adopting a molecular docking technology, the affinity experiment determines that the tripterine has relatively strong affinity with the S protein, and the tripterine has an obvious inhibition effect on the coronavirus in a cell experiment within a range of 5-40 mu M and has concentration dependence. The tripterine is a specific inhibitor of coronavirus and has specific novelty. The tripterine can be used for preparing the products for inhibiting the coronavirus and has wide practicability.
Owner:CAPITAL UNIVERSITY OF MEDICAL SCIENCES
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com