Molecular docking result screening method based on positive compound residue contribution similarity
A screening method and molecular docking technology, applied in the analysis of two-dimensional or three-dimensional molecular structure, molecular design, bioinformatics, etc., can solve problems such as low success rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
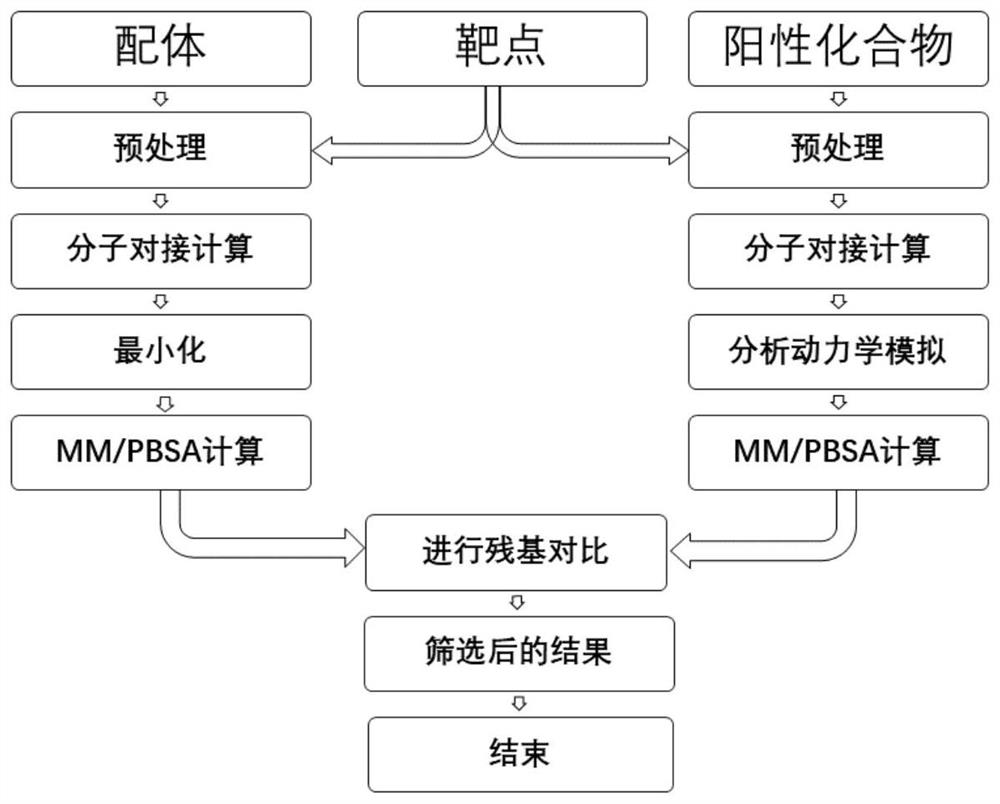
[0038] A molecular docking result screening method based on the contribution similarity of positive compound residues, the flow chart is shown in Figure 1, and the specific steps are as follows:
[0039] 1) Construction of a positive compound library for EGFR targets
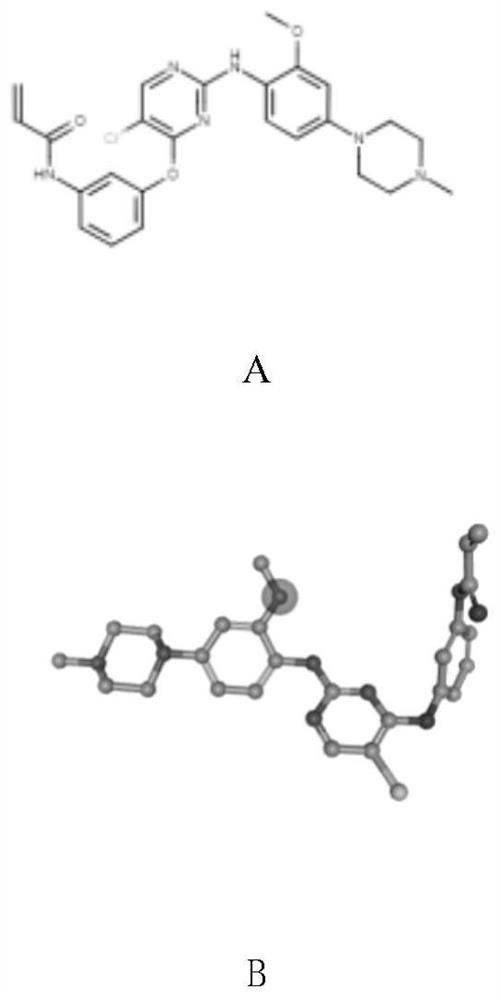
[0040] Obtain the crystal structure of the EGFR target from the protein database. The database also provides the binding force between the target and the ligand compound obtained by the Kd, IC50, Ki and other experimental methods of the crystal structure. The binding degree is selected as nanomolar and the experiment Compounds with small values. Use molecular visualization tools to separate crystal structures and obtain structural information data for positive compounds, such as figure 2 and image 3 shown. According to the requirements of the docking software, use the format conversion tool to convert the obtained positive compound structure data into a format that meets the requirements.
[0041] 2) Struc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com