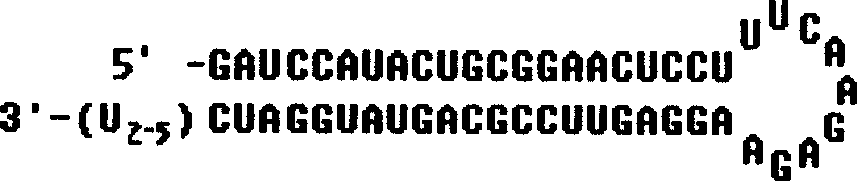Patents
Literature
161results about How to "Highly conservative" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
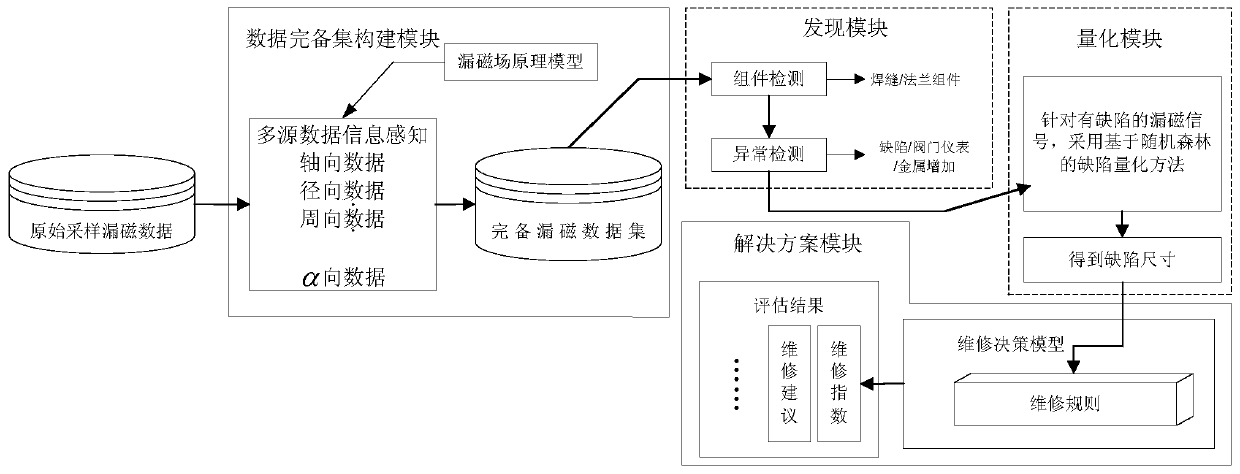
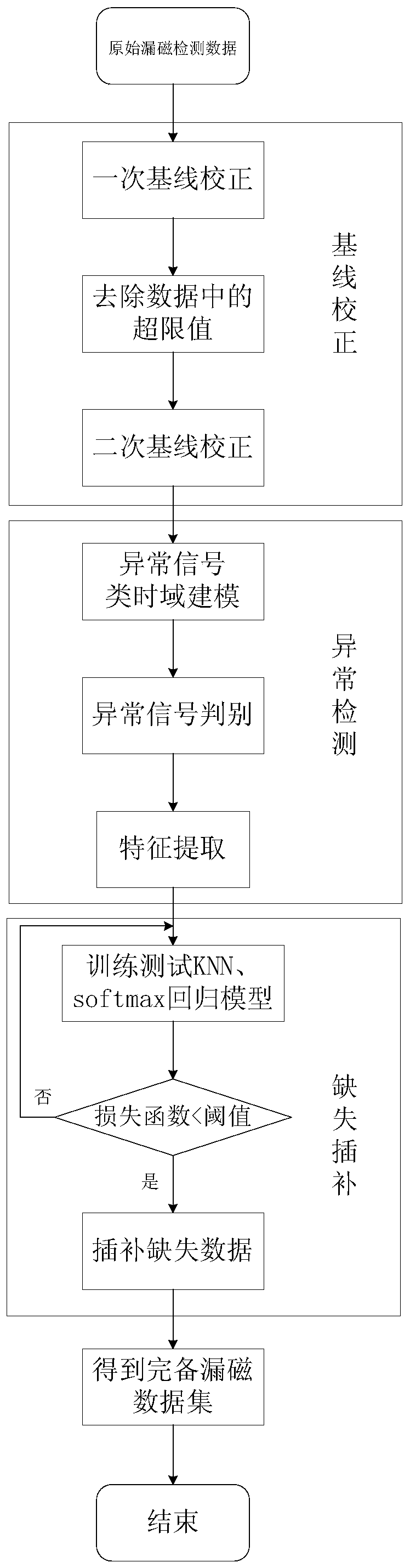
Intelligent analysis system and method for magnetic flux leakage detection data in pipeline
ActiveCN109783906AImprove accuracyImprove anti-interference abilityPipeline systemsNeural architecturesTime domainData set
The invention provides an intelligent analysis system and method for magnetic flux leakage detection data in a pipeline. The process comprises the following steps: adopting time domain-based sparse sampling and KNN-softmax data complete set construction module, obtaining a complete magnetic flux leakage data set; a pipeline connection assembly discovery method based on combination of selective search and a convolutional neural network is adopted in the discovery module to obtain the accurate position of the weld joint; adopting an abnormal candidate region searching and identifying method based on a Lagrangian multiplication frame and multi-source magnetic flux leakage data fusion in the discovery model to find out defective magnetic flux leakage signals; Adopting a defect quantification method based on a random forest in a quantification module to obtain a defect size; a pipeline solution improved based on the ASME B31G standard is adopted in the solution module, and an evaluation result is output. An analysis method is provided from the overall perspective, and pretreatment, connection assembly detection and abnormity detection, defect size inversion and final maintenance decision are achieved.
Owner:NORTHEASTERN UNIV
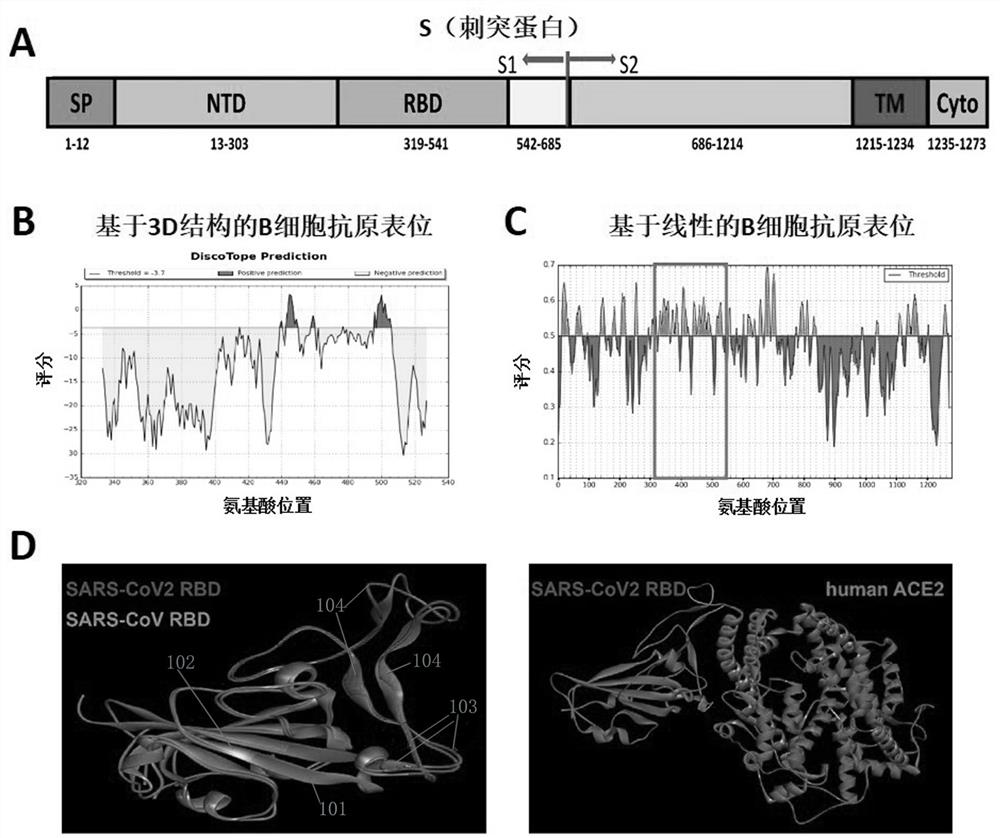
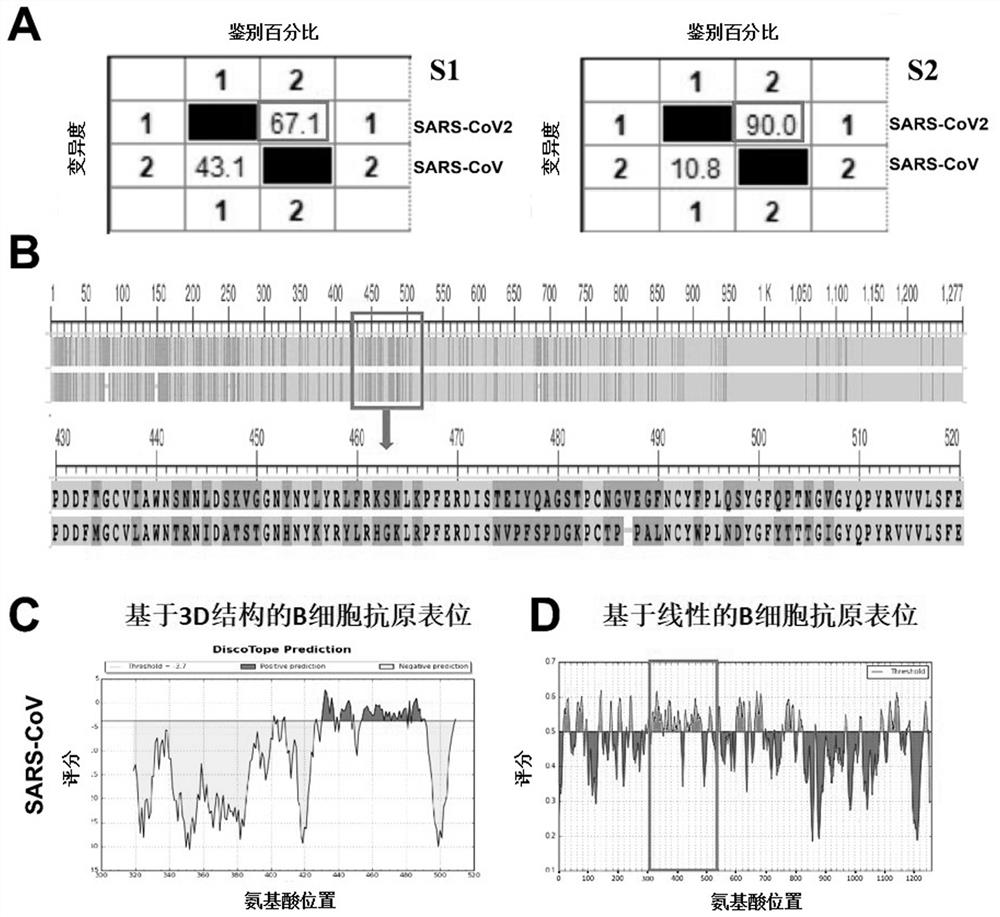
Novel coronavirus antigen epitope and application thereof
ActiveCN111848753AImproving immunogenicityHigh titerSsRNA viruses positive-senseAntibody mimetics/scaffoldsAntigen epitopeCoronavirus vaccination
The invention provides a novel coronavirus antigen epitope and application thereof. A super computer is used for simulating S, E, M and N protein structures of the novel coronavirus, and novel coronavirus epitopes SEQ ID NO 1-38 are obtained through calculation. The epitopes have very good immunogenicity; an antibody generated by inducing a part of epitope polypeptide has the effect of neutralizing the novel coronavirus; the antigen epitope can be combined with antibodies in serum of novel coronavirus patients at home and abroad, has the potential of resisting various novel coronavirus strains, and also has the potential of identifying and applying different strains. The epitope provided by the invention can be used for (1) research and development of novel coronavirus vaccines and universal coronavirus vaccines such as MERS viruses and SARS viruses; and (2) preparing a novel coronavirus antibody, and further detecting and typing viruses and treating diseases caused by the viruses. Thenovel coronavirus antigen epitope has a wide application prospect in the aspects of prevention of coronavirus and propagation and outbreak thereof, and detection and diagnosis of virus strains.
Owner:INST OF PROCESS ENG CHINESE ACAD OF SCI
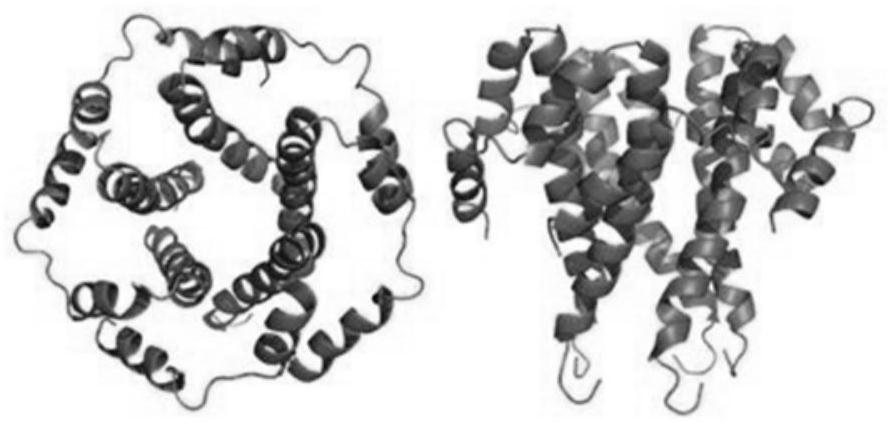
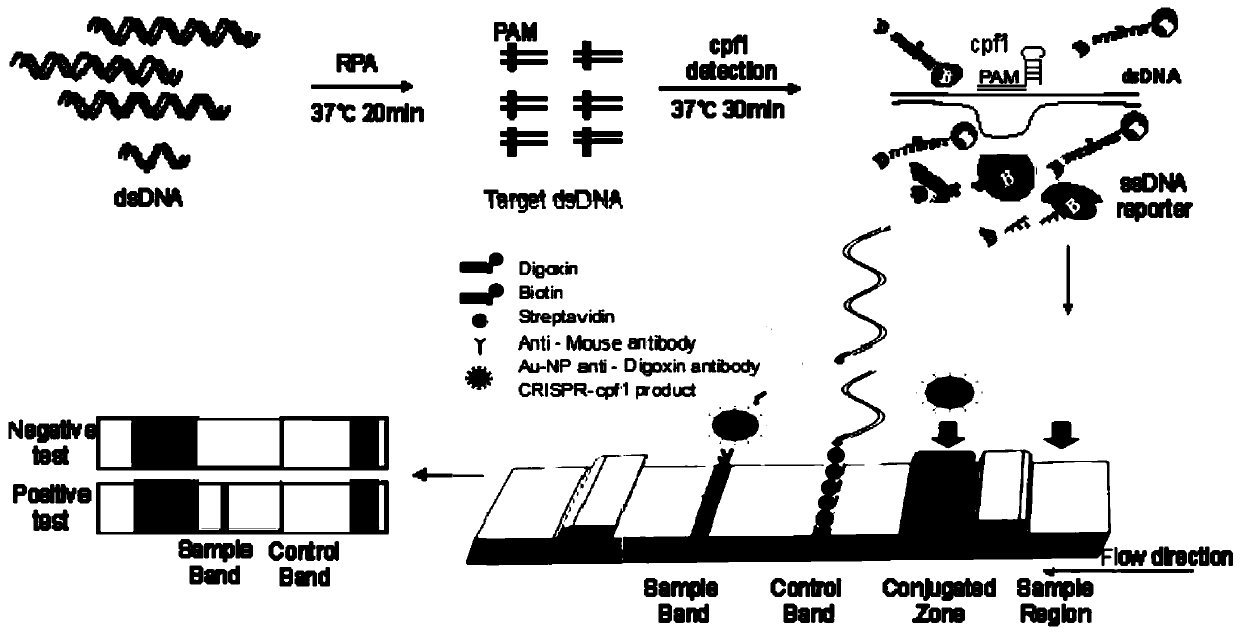
Cpf1 reagent kit and detection method for quickly detecting nucleic acid of African swine fever virus
ActiveCN110551846AHigh sensitivityStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationAfrican swine feverFluorescence
The invention discloses a Cpf1 reagent kit for quickly detecting nucleic acid of an African swine fever virus. The Cpf1 reagent kit comprises a Cpf1 detection system suitable for quickly detecting theAfrican swine fever virus, and an immune colloidal gold test strip, wherein the Cpf1 detection system comprises specific crRNA protein, specific Cpf1 protein and a single-chain DNA(ssDNA) reporting system in accordance with a p72 gene of the African swine fever virus, the specific crRNA is one or more of crRNAs from ASFV P72 crRNA1 to ASFV P72 crRNA10, and the sequence of the specific crRNA is SEQ NO.4 to SEQ NO.13; and the single-chain DNA(ssDNA) reporting system comprises ssDNA FQreporter for fluorescence detection of a microplate reader and / or ssDNA DB reporter for detecting the immune colloidal gold test strip. According to the Cpf1 reagent kit disclosed by the invention, for the first time, the Cpf1 is used for detecting the African swine fever virus, and has the advantages of beinghigh in sensitivity, high in specificity, short in time consumption, high in flux, independent of large-scale experiment equipment and the like. The advantages enable a detection method based on the immune colloidal gold test strip developed by the invention to be conveniently used in basic laboratories and breeding enterprises to be used for performing detection, identification and diagnosis on basic quick detection of the African swine fever.
Owner:SHANGHAI TECH UNIV
Novel vaccine for preventing COVID-19 and preparation method thereof
ActiveCN111939250AHighly conservativeAntibody induction ability is weakSsRNA viruses positive-senseAntibody mimetics/scaffoldsNucleotideReceptor
Provided is a novel vaccine for preventing COVID-19, the nucleotide sequence of an antigen of the novel vaccine is SEQ NO: 1, the amino acid sequence of the antigen of the novel vaccine is SEQ NO: 2,and the antigen of the vaccine comprises two functional parts: an S protein receptor binding structural domain capable of inducing a specific neutralizing antibody and a T cell related N protein truncated peptide fragment capable of inducing and activating effector T cells; The vaccine disclosed by the invention has the characteristics that the T cell related N protein truncated peptide fragment has weak capability of inducing the generation of the N protein antibody, so that a vaccine inoculator and a COVID-19 infected patient can be identified by using the N protein antibody, and the vaccineantigen does not induce the generation of the N protein antibody, so that lung injuries can be reduced, and the vaccine is safer. The cell vaccine disclosed by the invention is low in manufacturing cost, and can induce generation of virus-specific neutralizing antibodies and T cell immune response.
Owner:ZHENGZHOU UNIV
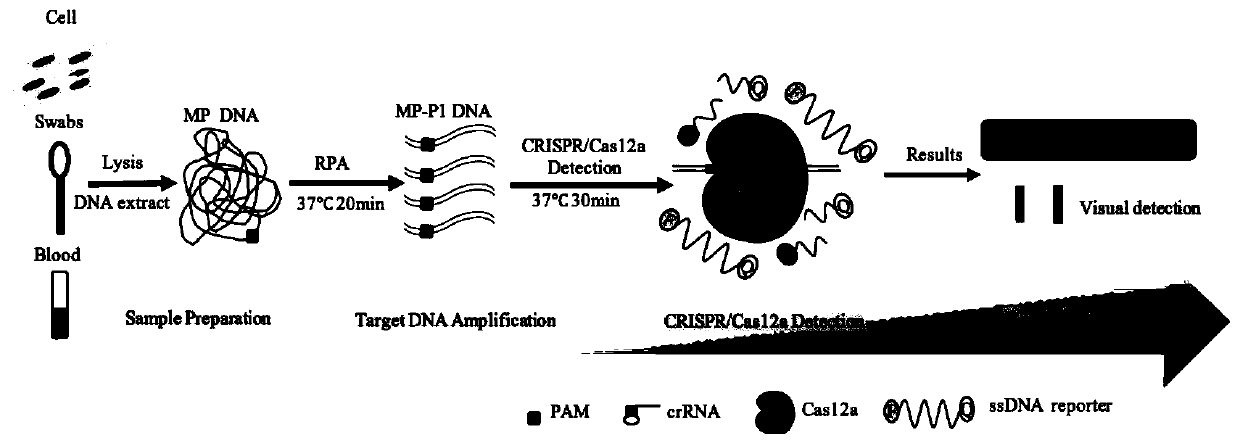
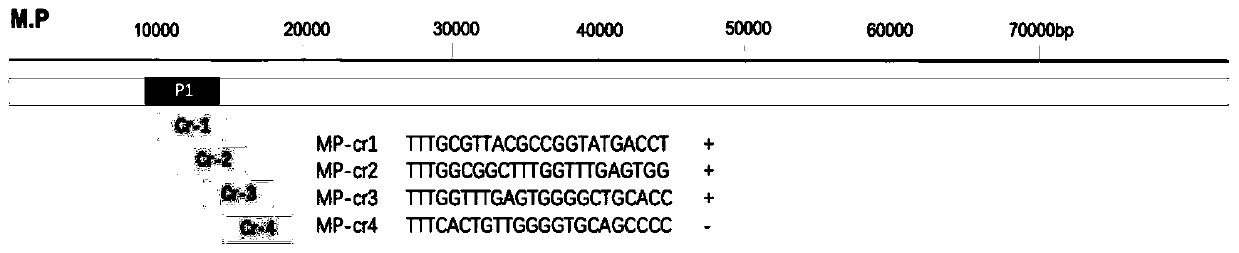
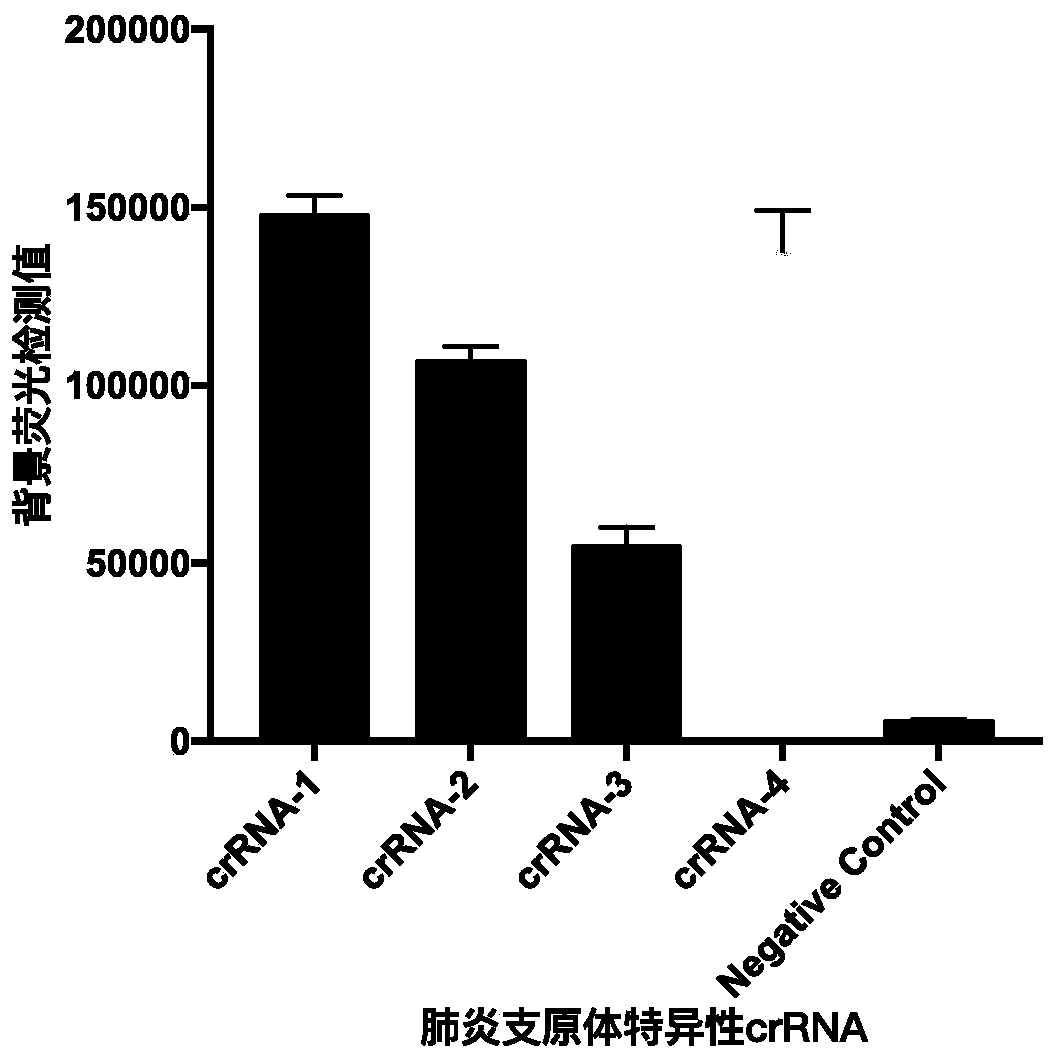
Kit and detection method for rapid detection of nucleic acid of mycoplasma pneumonia on basis of CRISPR/Cas12a
InactiveCN111187804AHigh sensitivityHighly conservativeMicrobiological testing/measurementMicroorganism based processesSingle strandNucleic acid sequencing
Owner:国家卫生健康委科学技术研究所
Polypeptide, immunogenicity conjugate and influenza vaccine
ActiveCN107488218AImprove survival rateFight infectionSsRNA viruses negative-senseViral antigen ingredientsAntigenImmune effects
The invention discloses a polypeptide, an immunogenicity conjugate and an influenza vaccine, and relates to the bio-technical field. The polypeptide is any one of a), b) or c): a) a polypeptide with the amino acid sequence as shown in SEQ ID NO.1; b) fused protein obtained by connecting a label to the end N and / or the end C of the polypeptide with the amino acid sequence as shown in SEQ ID NO.1; c) a polypeptide which is obtained by substitution and / or deletion and / or adding of one or several amino acid residues for the amino acid sequence as shown in SEQ ID NO.1 and has the same function. The polypeptide can be used for preventing or treating influenza virus, and can be used as an active component for preparing a medicine for preventing or treating the influenza virus. The immunogenicity conjugate disclosed by the invention can improve the immunogenicity and the immune effect on an antigen at a conformational dependency site.
Owner:华兰生物疫苗股份有限公司
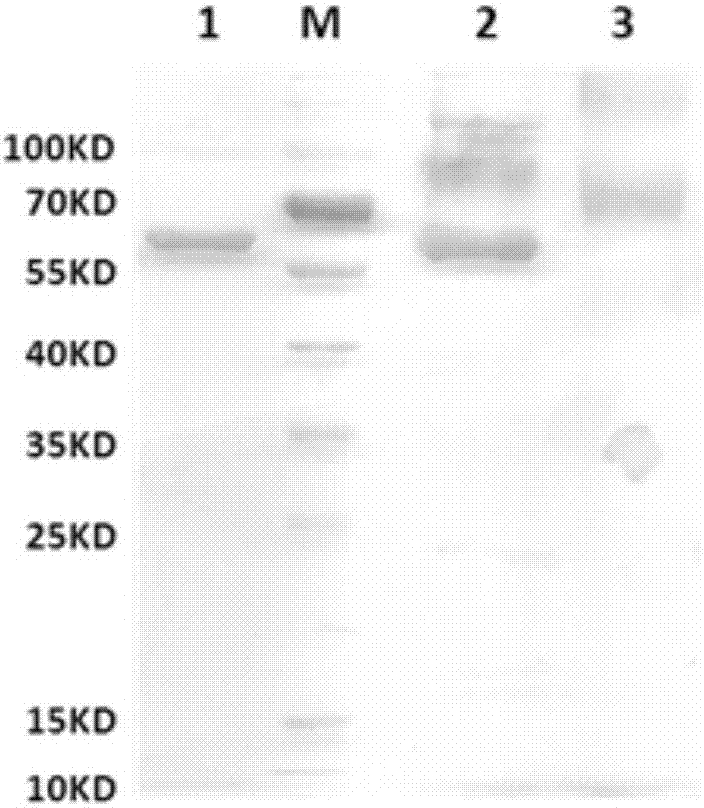
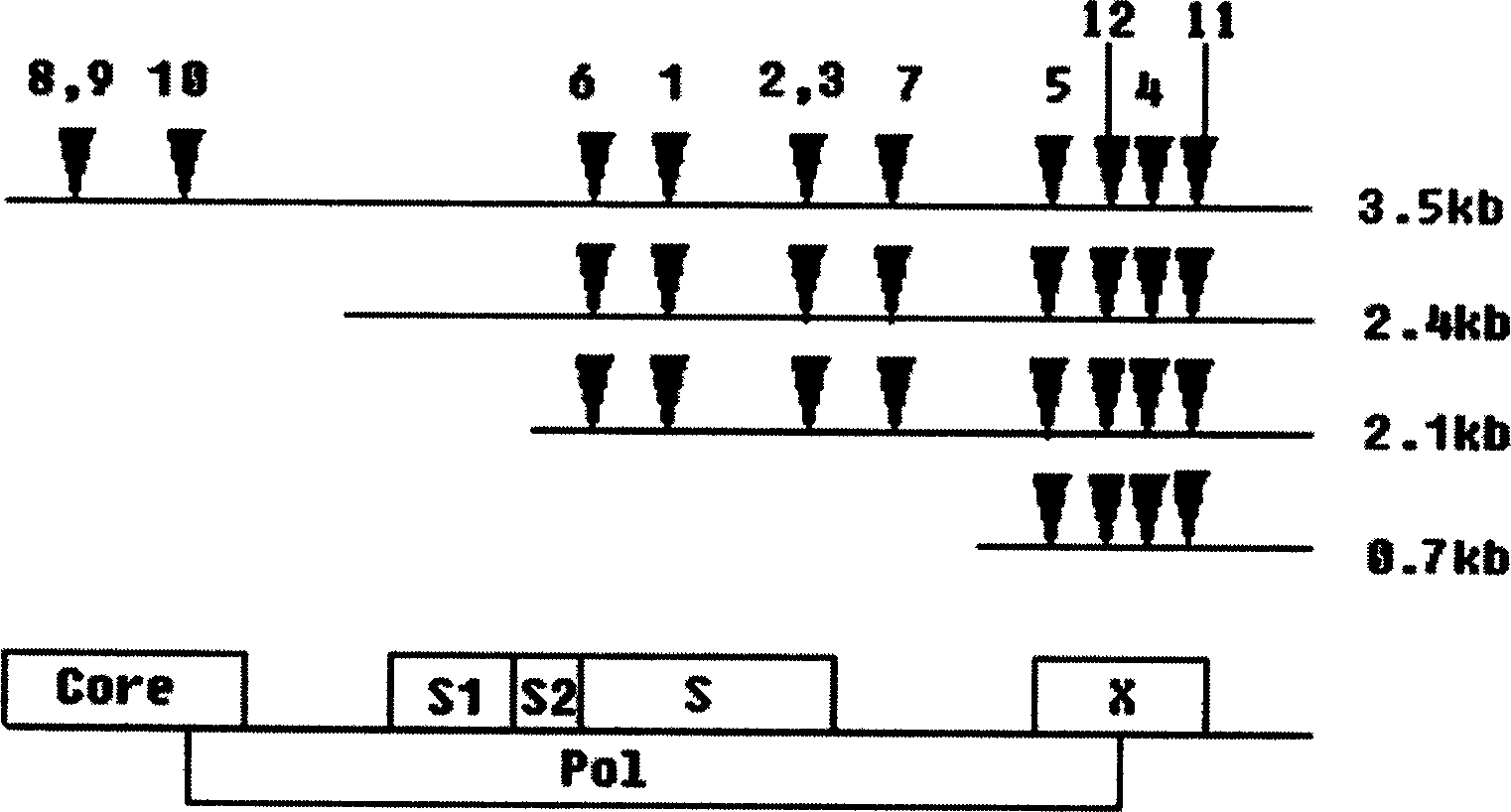
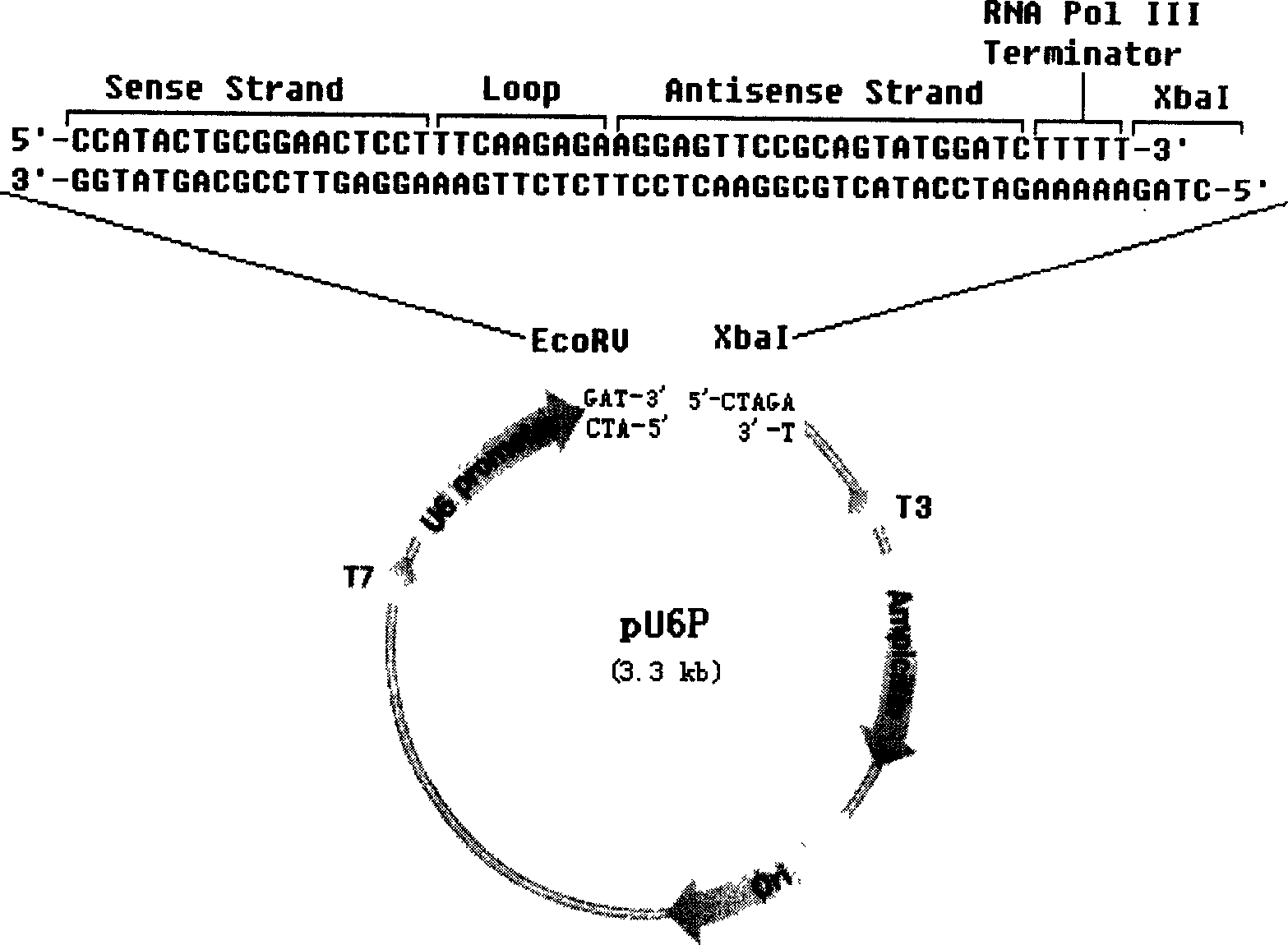
RNA interfered target sequence of HBV and use thereof
InactiveCN1667120AHighly conservativeNo homologyGenetic material ingredientsDigestive systemVirus ProteinIn vivo
This invention belongs to molecule biology and biomedicine technique field. It relates to 9 target points of RNA disturbing to HBV and applications of preparing hepatitis B medicine according to these target sequence. Plasmid is expressed through shRNA and siRNA transfection cell and animal cell is chemical synthesized. Then 16 RNA target point on HBV tissue is sieved, 9 of them can obvious restrain HBV protein expression at cell level. Expression and virus duplication of HBV protein inside cell can be stably restrained by 11 target point, and expression of HBV transgene little mouse in vivo virus protein can be restrained. Medicine curing hepatitis B can be made according to these target sequence.
Owner:SUN YAT SEN UNIV
Method for developing functional molecular marker related to miRNA
InactiveCN102222175AImprove stabilityHigh polymorphismSpecial data processing applicationsAmplified fragment length polymorphismBase-Base Mismatch
The invention discloses a method for developing a functional marker related to MicroRNA (miRNA for short). The method comprises the following steps of: performing BlastN comparison on a known miRNA sequence in an miRNA database with a disclosed DNA sequence of a researched species; determining a DNA (miDNA for short) sequence corresponding to the miRNA based on the number and a similarity degree of base mismatch; extending to the left and right for a certain distance until the total length is about 150bp; determining a precursor miDNA site through prediction of a stem loop secondary structure and calculation of the minimum free folding energy coefficient; classifying and comparing all screened precursor miRNA sequences; designing primers according to conserved sequences; electronically comparing the designed primers in the database, and determining as miRNA special sequences; and finally combining according to an annealing temperature of each primer, and developing the functional marker with high efficiency and good polymorphism. The marker system integrates the advantages of the conventional markers such as simple sequence repeat (SSR), amplified fragment length polymorphism (AFLP) and the like, and has good characteristics of higher portability, low cost, polymorphism and stability and the like.
Owner:SOUTHWEST UNIVERSITY
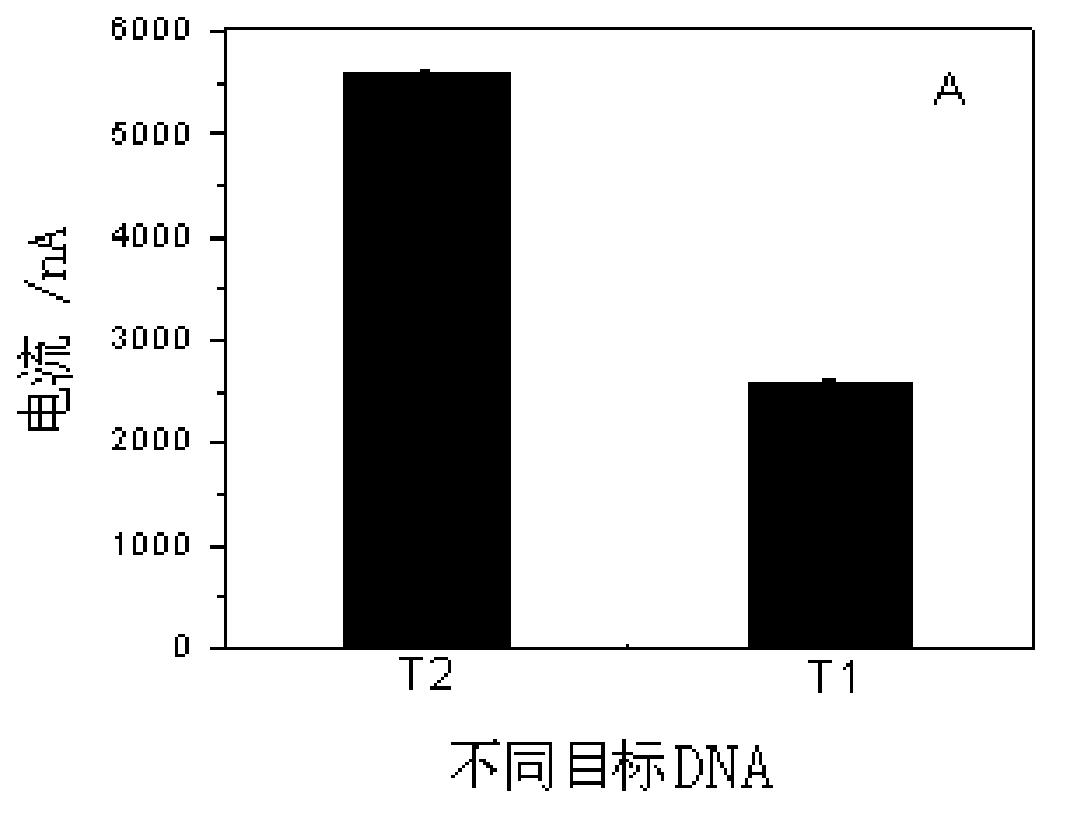
DNA electrochemical sensor based on target DNA repetitive sequence self enhancement and amplification signal
InactiveCN103175873ARealize highly sensitive detectionHighly conservativeMaterial electrochemical variablesRepetitive SequencesPower flow
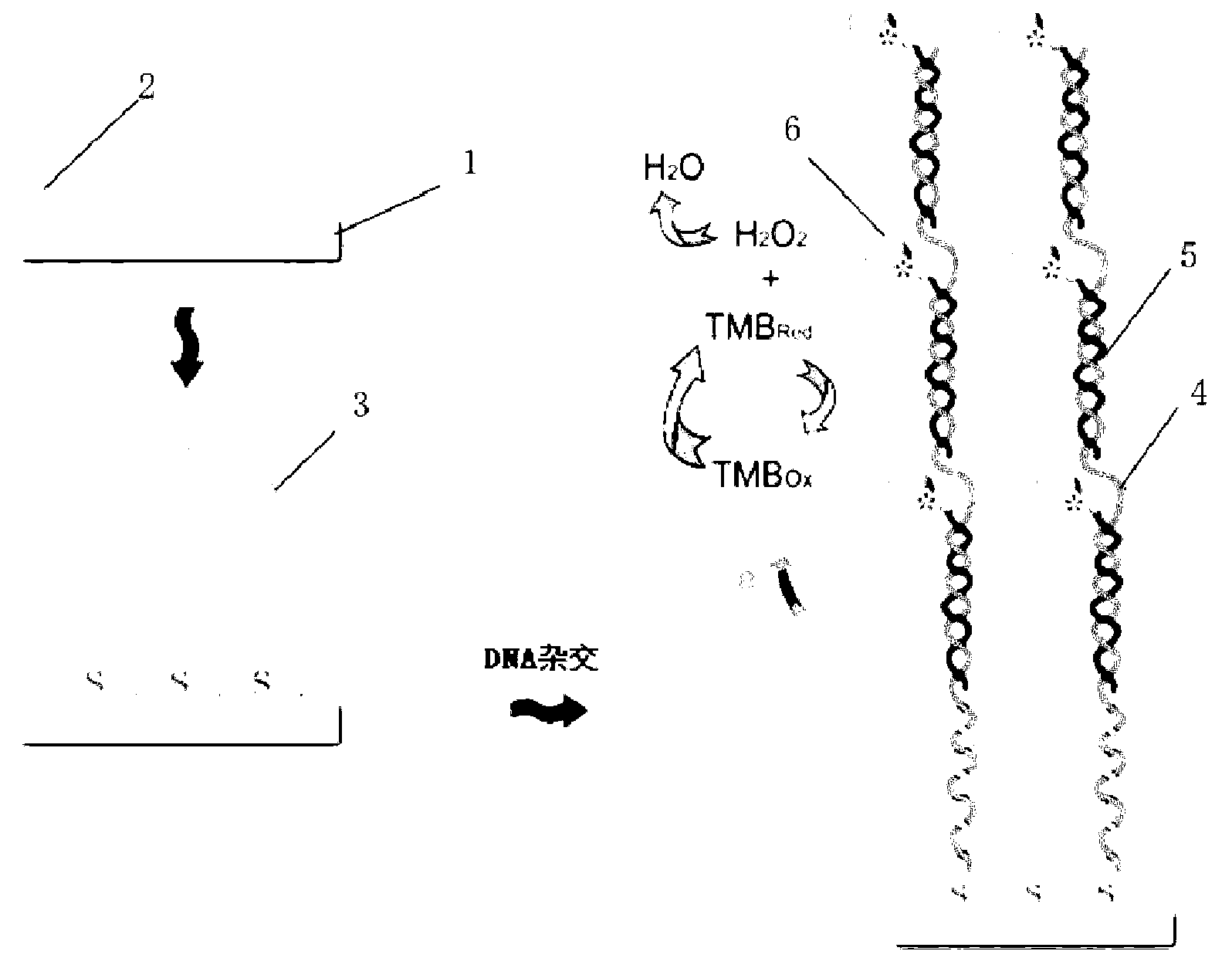
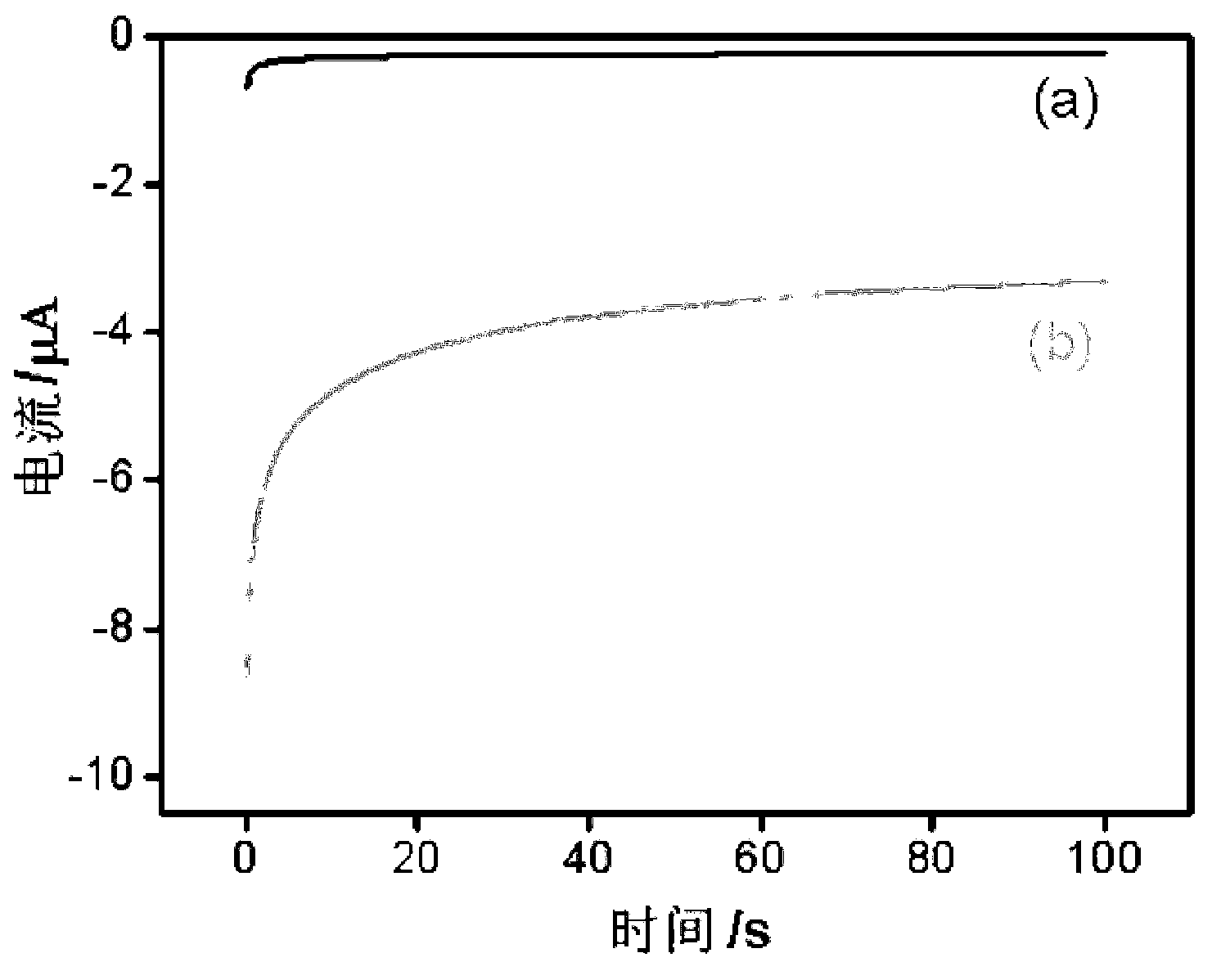
The invention discloses a DNA electrochemical sensor based on target DNA repetitive sequence self enhancement and amplification signal. A signal probe is designed by a repetitive sequence clip of a detected target chain, thus, the volume of the signal probes combined with the target sequence is increased, and a compound is formed, when a capture probe on electrode surface is hybridized with the target sequence, the amplified signal probe is combined with the electrode surface, under the affinity function between the biotin treated by end labeling of the signal probe and the avidin modified on the horseradish peroxidase, multiple enzymes are combined with a 'sandwich' structure, finally, under the efficient catalytic action of the enzymes, H2O2 is catalyzed to oxidize the TMB, the oxidative product generates an amplified reduction current signal on the electrode, and the concentration of the target DNA is judged according to the magnitude of the current signal. In the experiment, based on the characteristics of self enhancement and amplification signal of the target chain of the repetitive sequence, the target chain with two sections of repetitive sequences is detected, and the method is high in sensitivity.
Owner:福州市第二医院 +1
PEDV S gene major antigen epitope serial connection recombination gene, and preparation method and application thereof
ActiveCN105925597ARapid diagnosisMonitoring and Assessing ProtectionSsRNA viruses positive-senseVirus peptidesAntigen epitopeImmunogenicity
The invention discloses a PEDV S gene major antigen epitope serial connection recombination gene, and a preparation method and an application thereof, and belongs to the field of a PEDV S protein major neutralizing epitope region efficient expression method. The PEDV S gene major antigen epitope serial connection recombination gene is obtained through connecting three major antigen epitopes of a PEDV S gene in series; every two major antigen epitopes are connected through a flexible amino acid base sequence; the name is S123; and the sequence of the base sequence is shown as SEQ ID NO:1. The recombination gene contains most PEDV antigen epitopes; the selected fragments avoid a high-variation region; the expression quantity is efficient and stable; the immunogenicity is high; when the application of PEDV S gene major antigen epitope serial connection recombination protein to a PEDV indirect ELISA detection kit is used, the PEDV can be fast diagnosed; and the swinery neutralization protection can be well monitored and evaluated.
Owner:SOUTH CHINA AGRI UNIV
Primer probe set, kit and detection method for detecting SARS-CoV (Severe Acute Respiratory Syndrome Coronavirus) and MERS-CoV (Middle East Respiratory Syndrome Coronavirus)
InactiveCN108060266AQuick judgmentShort timeMicrobiological testing/measurementAgainst vector-borne diseasesMiddle East respiratory syndrome coronavirusRecombinase Polymerase Amplification
The invention discloses a primer probe set, kit and detection method for detecting SARS-CoV and MERS-CoV based on Recombinase Polymerase Amplification (RPA) detection. The primer probe set for detecting SARS-CoV and MERS-CoV based on the RPA detection comprises primers for nucleotide sequences as shown in SEQ ID NO.1-4 and probes for nucleotide sequences as shown in SEQ ID NO. 5-6. The invention further provides the kit for detecting SARS-CoV and MERS-CoV based on the RPA detection, wherein the kit comprises the primer probe set. By adopting the technical scheme, the sensitivity, specificity and simplicity for detecting SARS-CoV and MERS-CoV are obviously improved.
Owner:北京卓诚惠生生物科技股份有限公司
Enzyme-linked immunosorbent assay vector and kit for detecting avian leukosis P27
The invention belongs to the technical field of animal epidemic disease serological diagnosis, and relates to an enzyme-linked immunosorbent assay vector and a kit for detecting avian leukosis P27. The avian leukemia virus (P27) enzyme linked immunosorbent assay kit comprises a 96 hole elisa plate coating a avian leukosis P27 polyclonal antibody, P27 monoclonal antibody marked by alkaline phosphatase, a substrate solution, a stop solution and a cleaning solution. The vian leukosis P27 polyclonal antibody and the P27 monoclonal antibody marked by alkaline phosphatase effectively improve the sensitivity, the specificity and the stability of the detection. The invention provides an efficient and sensitive ELISA (enzyme linked immunosorbent assay) detection kit for sifting out and purifying avian leukosis positive chickens and cultivating new species with heredity resistance, and the kit is low in cost and simple and convenient to operate, and is applicable to promotion in animal husbandry.
Owner:LANZHOU INST OF ANIMAL SCI & VETERINARY PHARMA OF CAAS +2
Method and kit for fast detection of moraxella catarrhalis based on magnetic resolution and quantum dot labelling
ActiveCN105203754AFast separationIt has the effect of synergistic amplification of multiple signalsFluorescence/phosphorescenceAntigenMagnetic bead
The invention provides a method for detection of moraxella catarrhalis based on magnetic resolution and quantum dot labelling. The method comprises the following steps: (1) preparing anti-moraxella catarrhalis nano immunomagnetic beads; (2) preparing quantum dot labelled anti-moraxella catarrhalis nanoprobes; (3) dissolving a sample to be detected with a PBST buffer solution, adding the anti-moraxella catarrhalis immune nano magnetic beads into the dissolved solution, carrying out magnetic separation after sufficient mixing and reaction, washing with the PBST buffer solution, adding the quantum dot labelled anti-moraxella catarrhalis nanoprobes into the obtained precipitate, carrying out magnetic separation after a reaction, washing with the PBST buffer solution, and detecting the fluorescence value with a fluorescence micro-plate reader. The method is accurate, fast, and high in sensitivity, and has very high practical value in the aspects of clinical diagnosis, etiological identification, epidemiological surveys and the like of moraxella catarrhalis.
Owner:湖北诺美华抗体药物技术有限公司
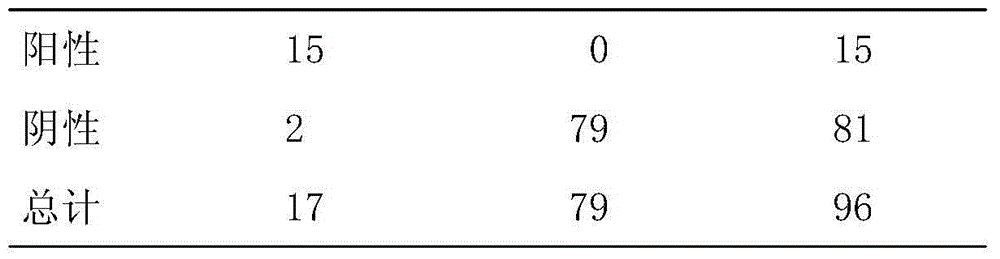
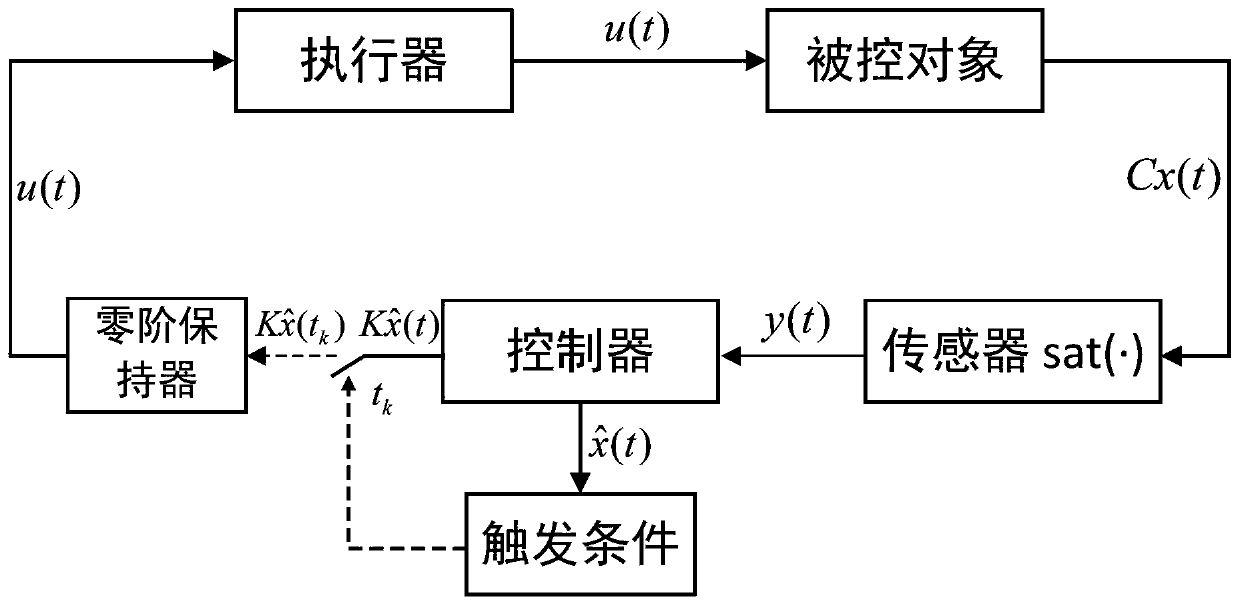
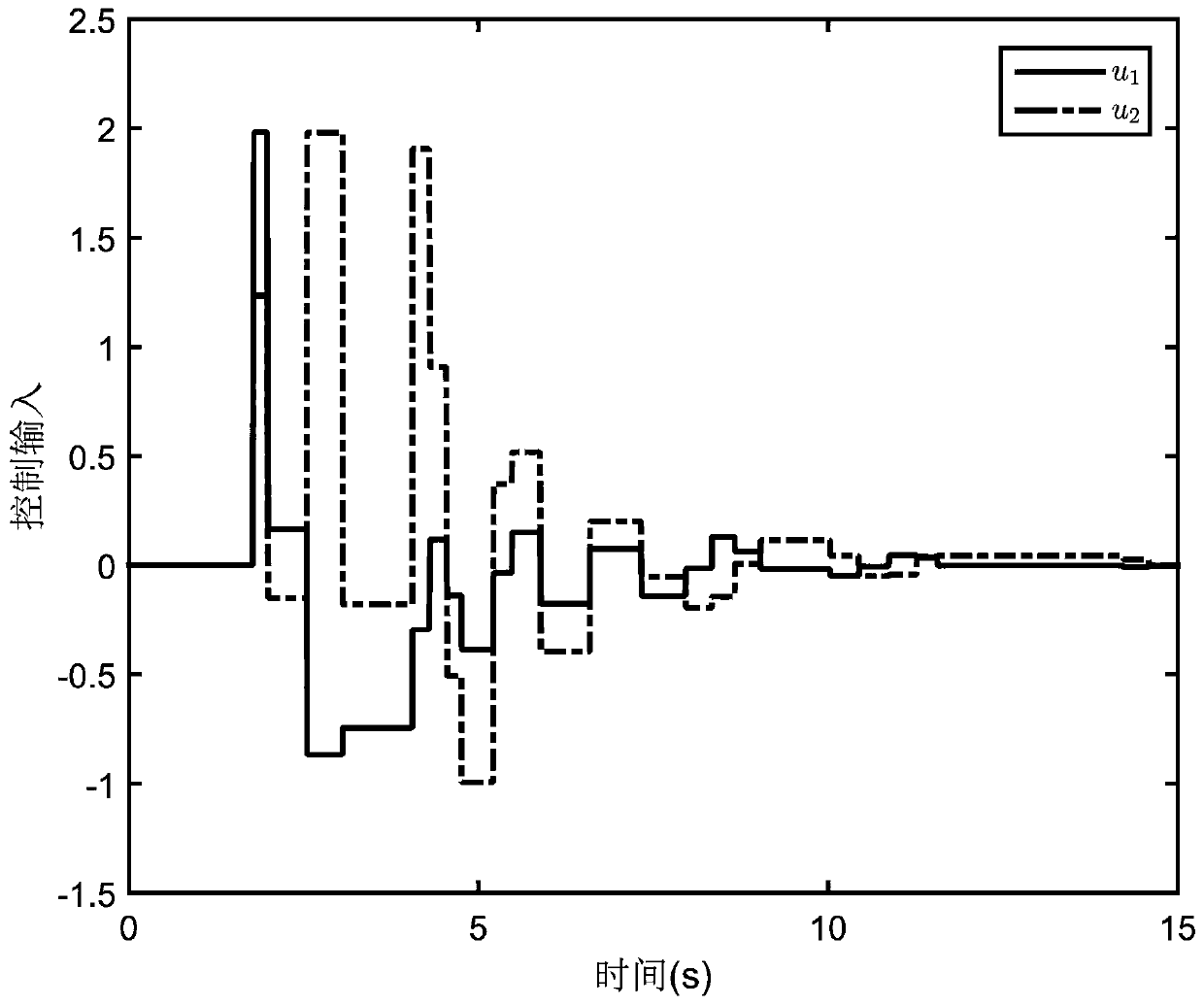
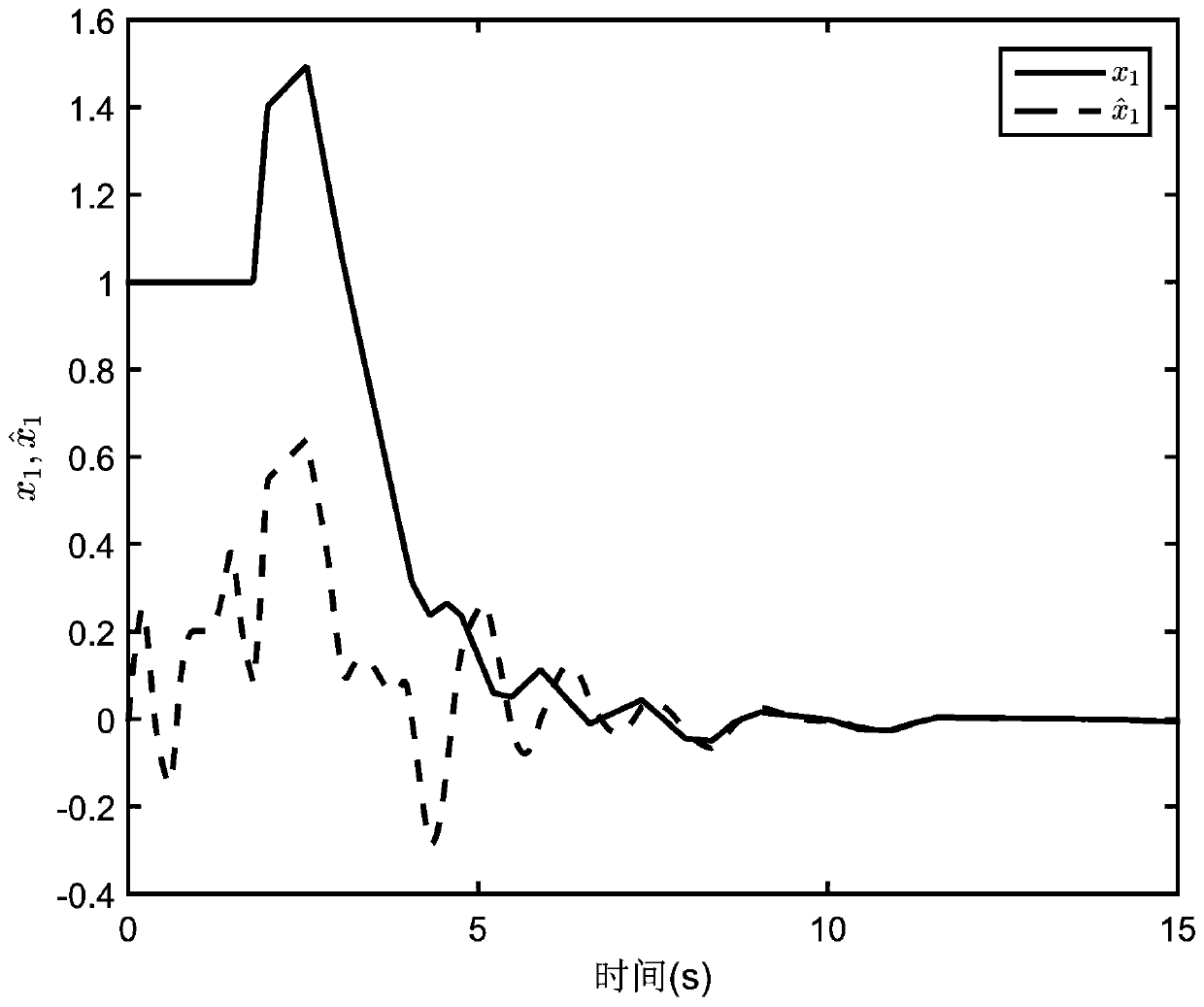
Event-trigger-based output feedback controller of neutral stable saturation system
ActiveCN110456681AAlleviate transmission burdenGuaranteed global asymptotic stabilityProgramme controlComputer controlMinimum timeEvent trigger
The invention relates to an event-trigger-based output feedback controller of a neutral stable saturation system. The design method comprises: establishing a model for a neutral stable linear system with output saturation characteristics; designing an output feedback control based on a state observer; determining a sampling time; providing models of an observer system and an observation error system under the event trigger action; providing stability conditions of the system; and providing a minimum time interval for event triggering to avoid the Zeno phenomenon in the control process.
Owner:TIANJIN UNIV
Nucleic acid reagent, kit, system and method for detecting invasive fungi
InactiveCN110551840AQuick checkAccurate detectionMicrobiological testing/measurementMicroorganism based processesCandida tropicalisCandida famata
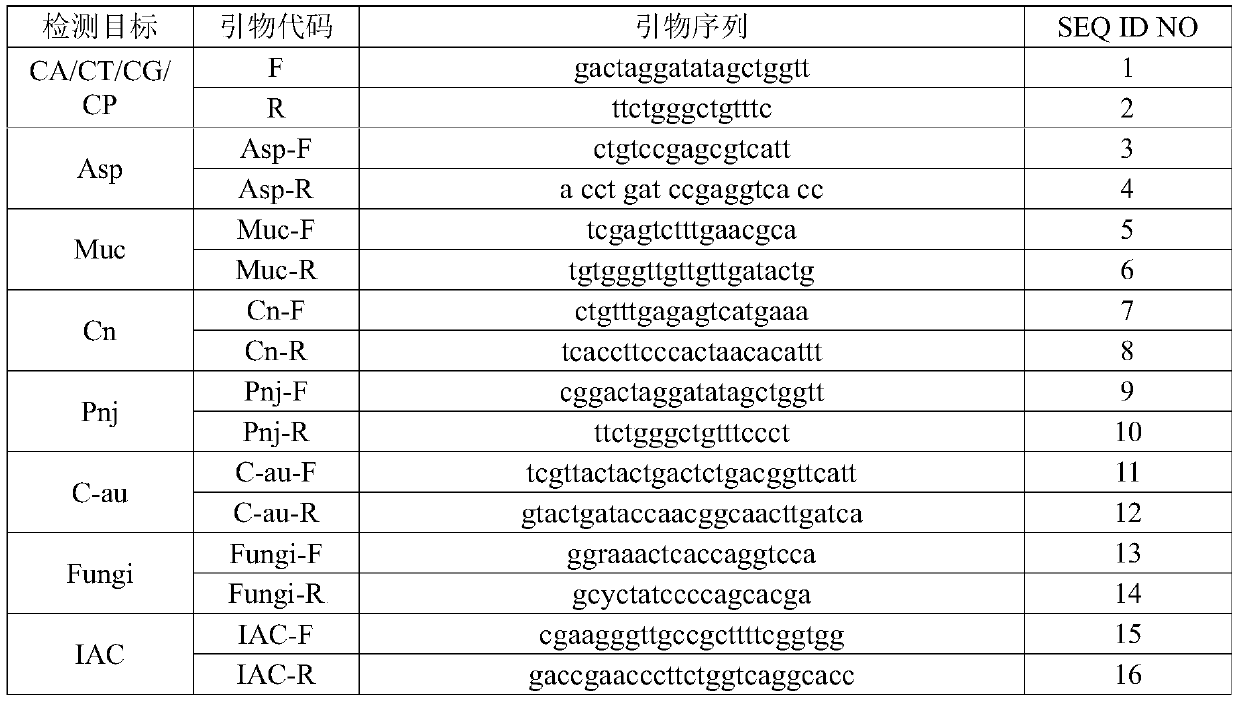
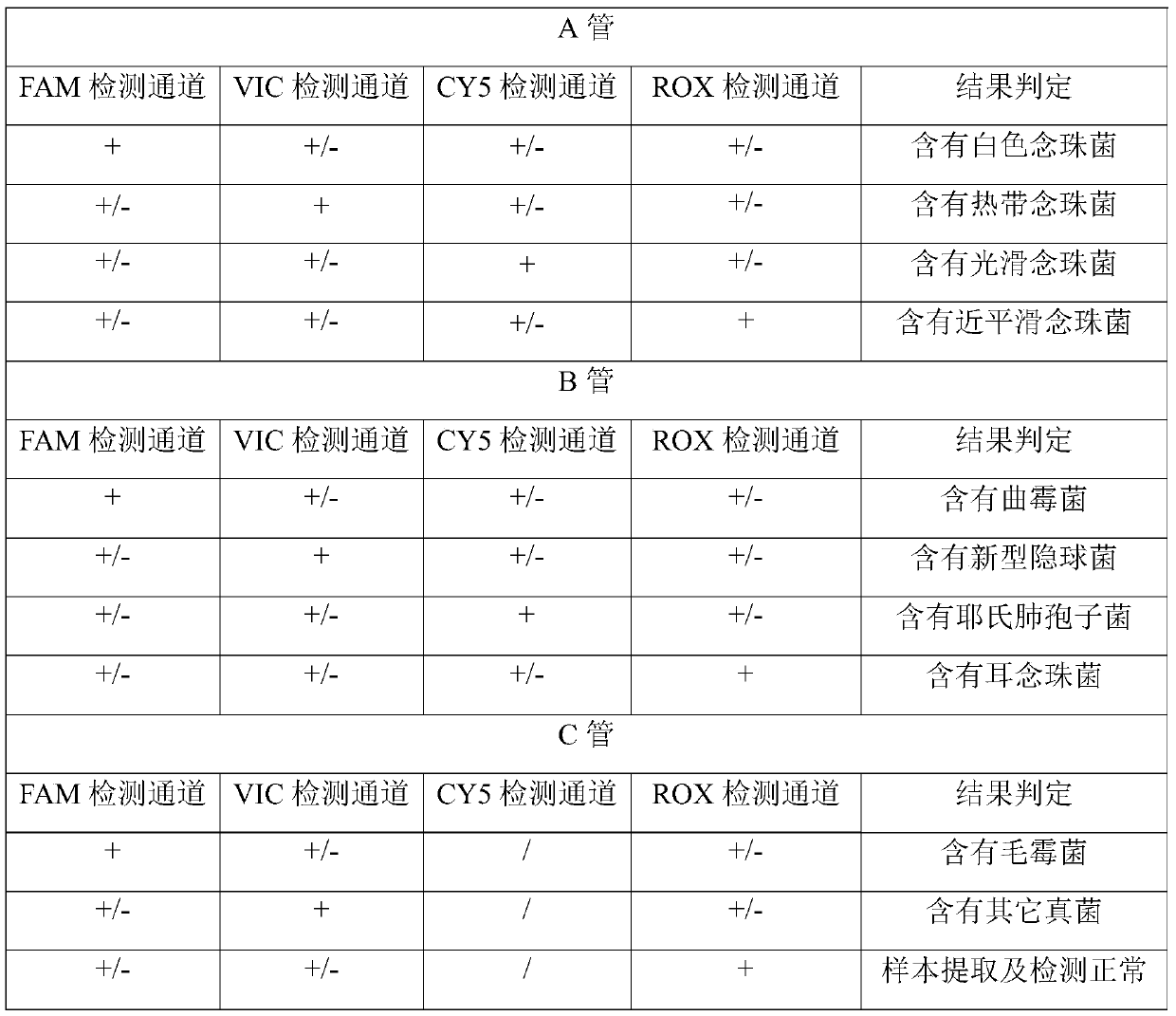
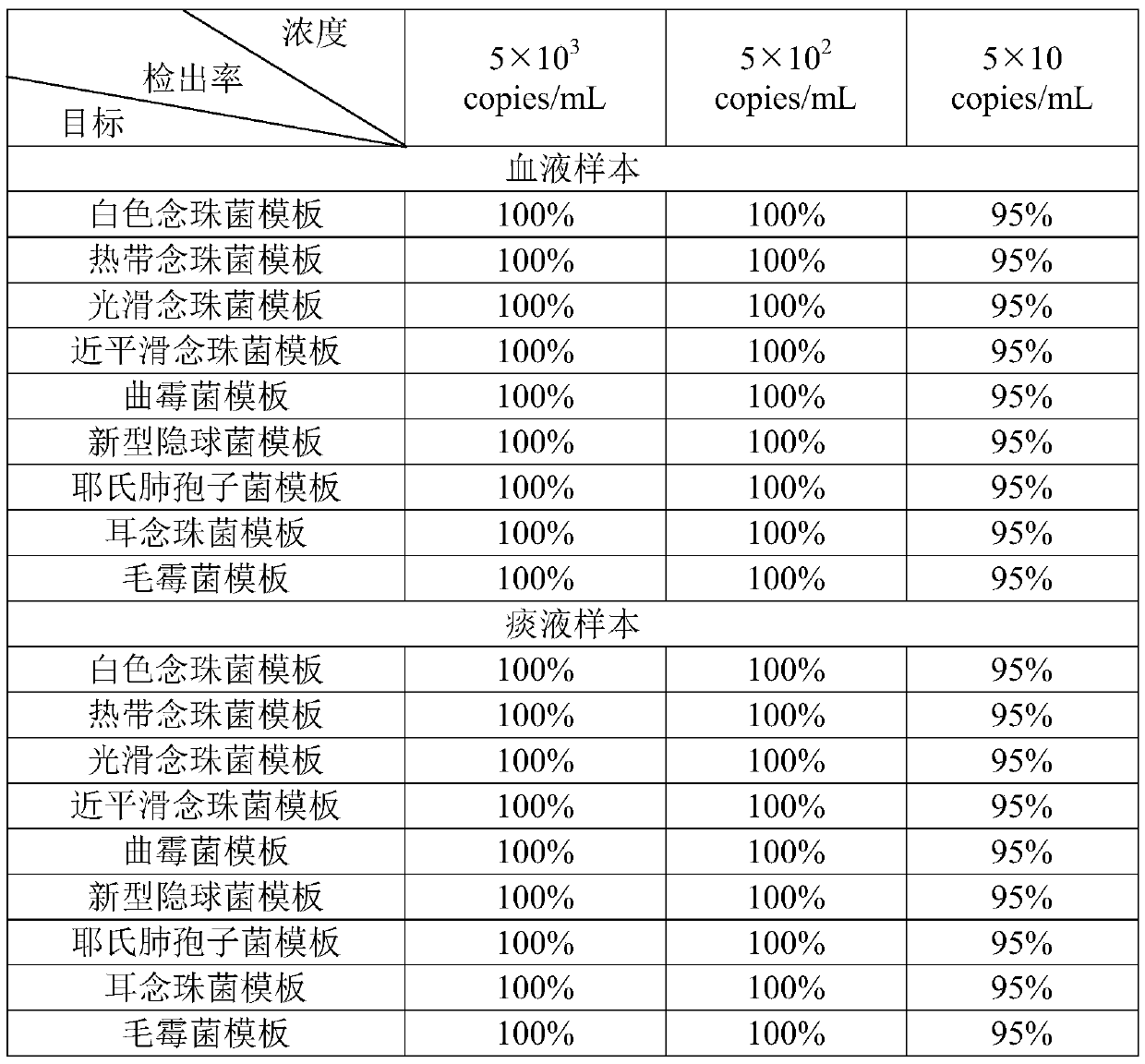
The invention relates to a nucleic acid reagent, kit, system and method for detecting invasive fungi. The nucleic acid reagent comprises a primer shown in SEQ ID NO.1-14 and a probe shown in SEQ ID NO.17-26, and the primer and the probe are stored independently with each other correspondingly or mixed with each other at will. Through the primer and the probe, the nucleic acid reagent, the kit, thesystem and the method of at least 9 invasive fungi of candida albicans, candida tropicalis, candida glabrata, candida parapsilosis, mucor, aspergillus, cryptococcus neoformans, pneumocystis jeroveciand candida auris, fast, comprehensive, sensitive, specific and automatic detection result determination can be achieved, and the sensitivity, specificity, simplicity and convenience of simultaneous detection of the detection target genome are significantly improved.
Owner:北京卓诚惠生生物科技股份有限公司
Nucleic acid identification sequence and detection method for singular proteus
InactiveCN101215567AHighly conservativeImprove featuresSugar derivativesMicrobiological testing/measurementNucleotideNucleic acid sequencing
The invention discloses a specific nucleic acid marking sequence of proteus mirabilis, a PCR detecting method and the application. The aim of the invention is to provide the specific nucleic acid marking sequence of the proteus mirabilis, a qualitative and quantitative PCR detecting method of the proteus mirabilis and the application in preparing qualitative and quantitative PCR detecting kits of the proteus mirabilis. The specific nucleic acid marking sequence of the proteus mirabilis is one of following nucleic acid sequences: firstly, a DNA sequence of SEQ ID NO: 1 in the table, secondly, a restricted DNA sequence of the SEQ ID NO: 1 in the table, which has the homology more than 90% and is the specific nucleic acid sequence of the proteus mirabilis, and thirdly, a nucleic acid sequence which can cross-breed with the restricted DNA sequence of the SEQ ID NO: 1 in the table under the high-stringent condition. The invention can be used in qualitative and quantitative molecular detection of the proteus mirabilis (including the detecting methods of ordinary PCR, quantitative PCR, chips, hybrid and the like) and has broad application prospect.
Owner:INST OF PLA FOR DISEASE CONTROL & PREVENTION
Process, computer-accessible medium and system for obtaining diagnosis, prognosis, risk evaluation, therapeutic and/or preventive control based on cancer hallmark automata
InactiveUS20120290278A1Highly conservativeMedical simulationAnalogue computers for chemical processesComputational modelHybrid automaton
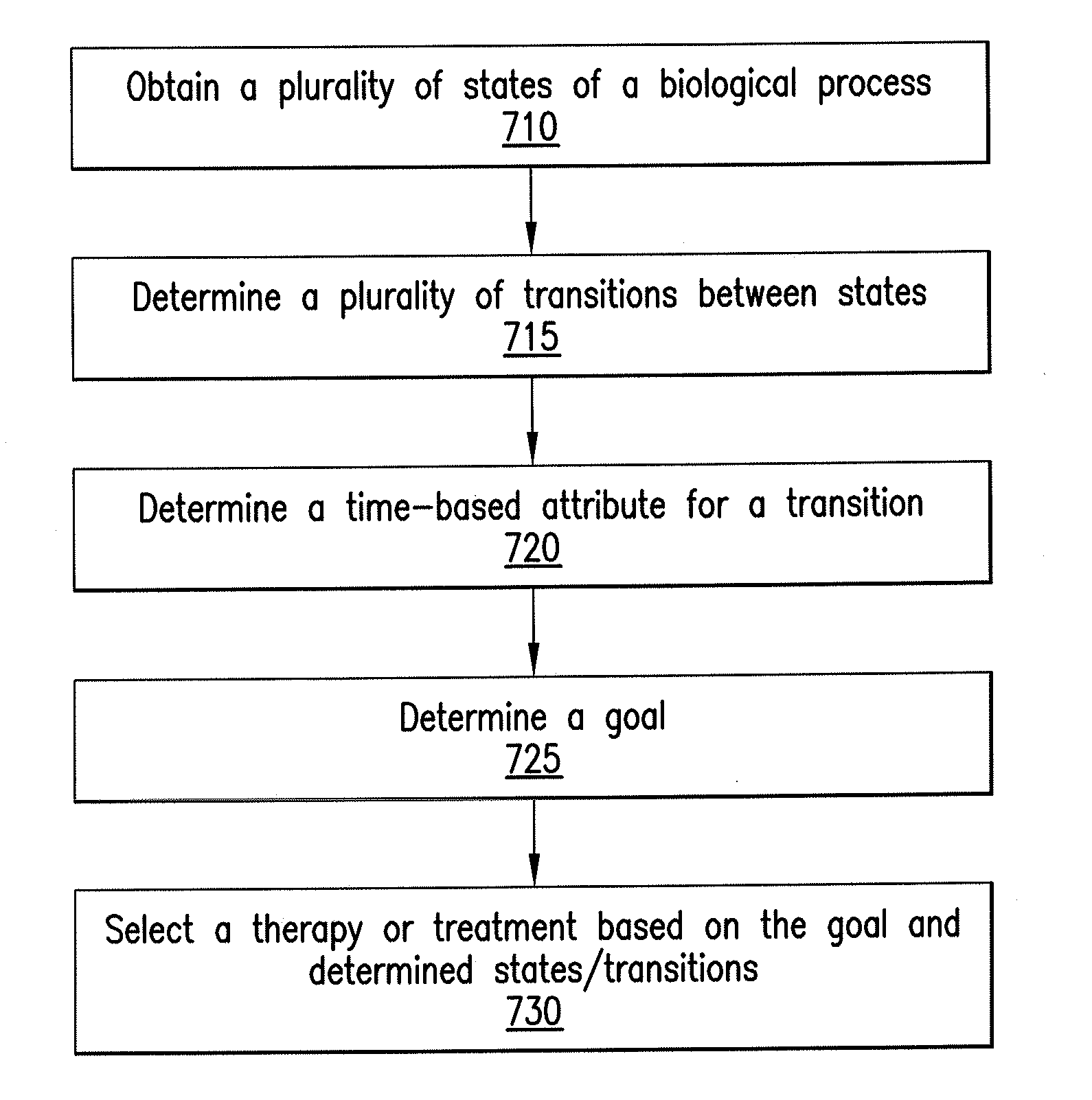
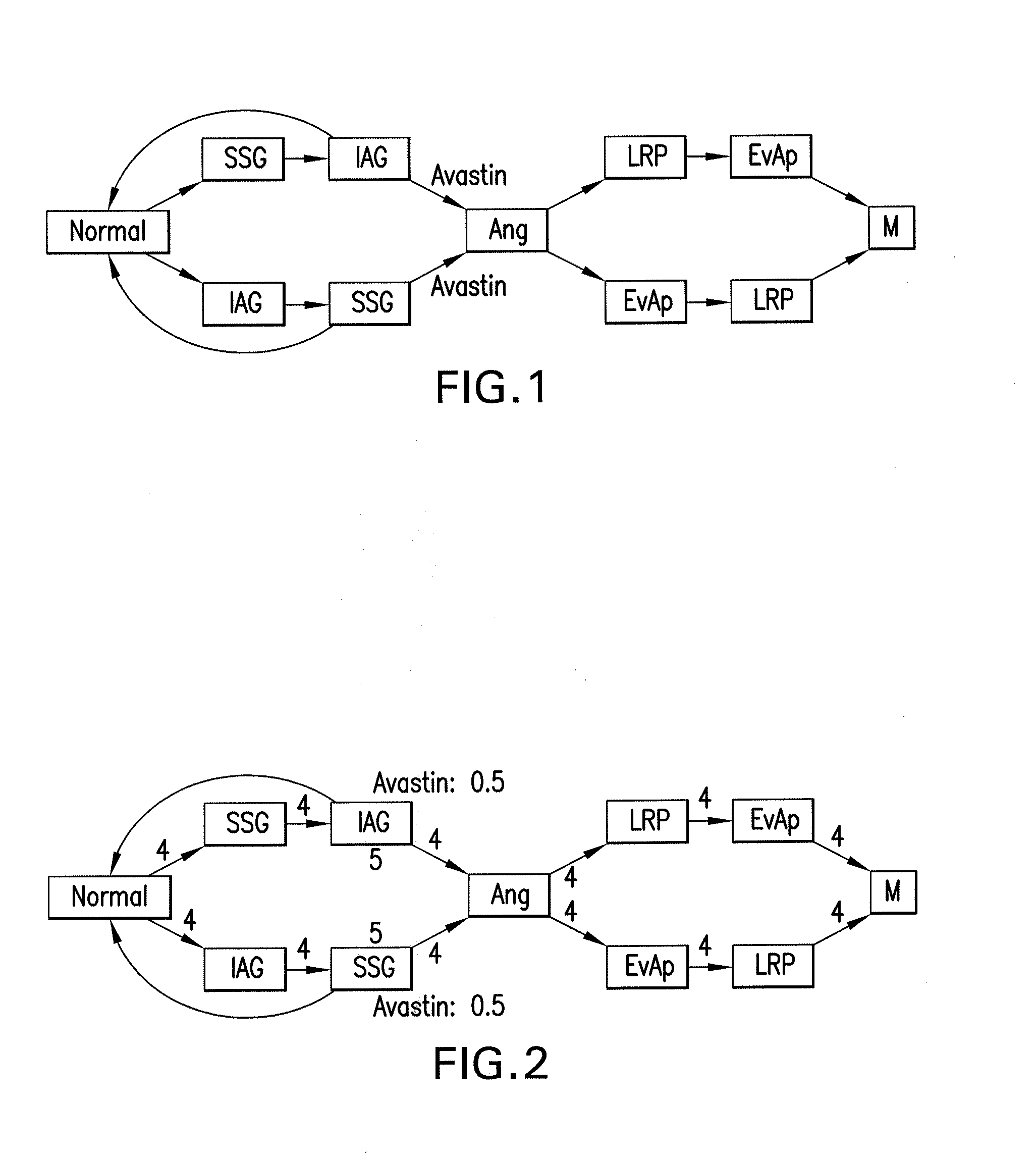
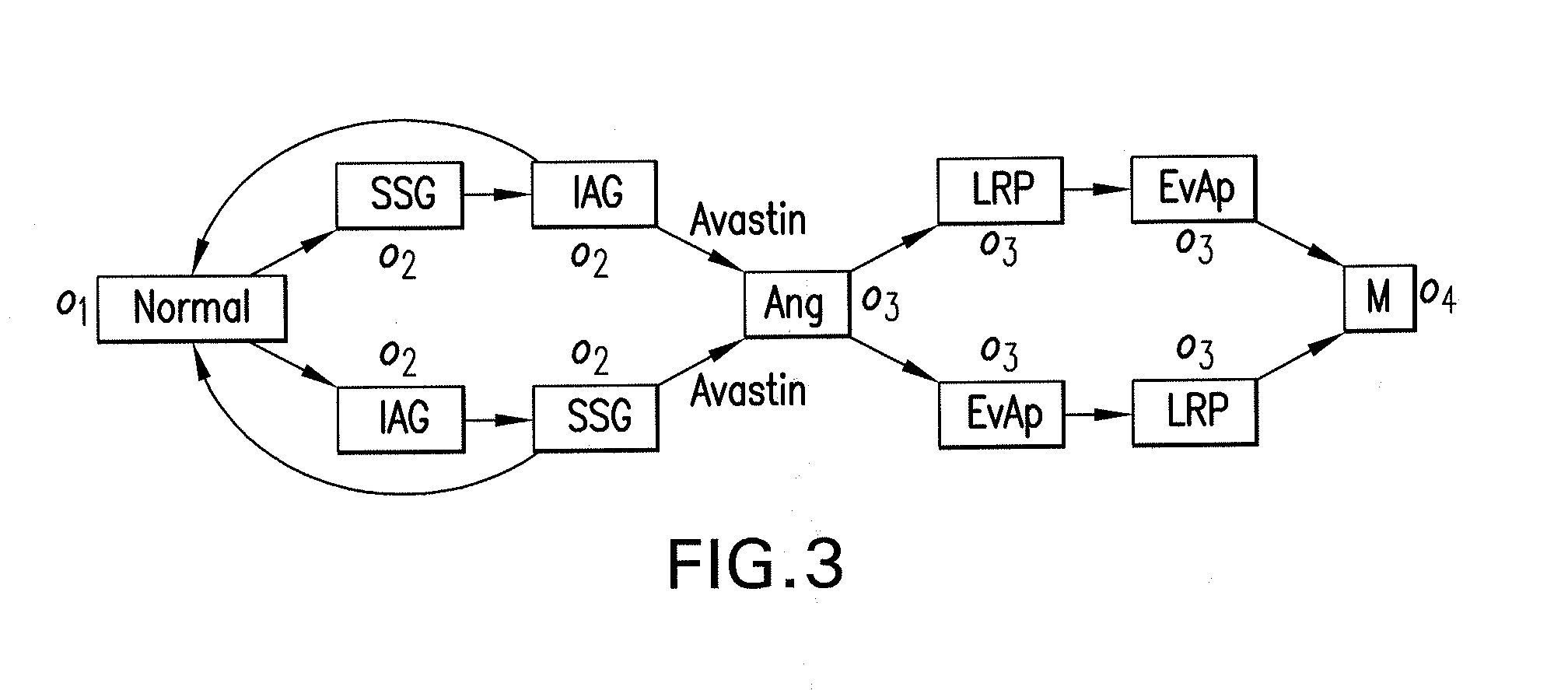
The present disclosure relates to exemplary embodiments of method, computer-accessible medium, system and software arrangements for, e.g., Cancer Hallmark Automata, a formalism to model the progression of cancers through discrete phenotypes (so-called hallmarks). The precise computational model described herein includes the automatic verification of progression models (e.g., consistency, causal connections, etc.), classification of unreachable or unstable states (e.g., “anti-hallmarks”) and computer-generated (individualized or universal) therapy plans. Exemplary embodiments abstractly model transition timings between hallmarks as well as the effects of drugs and clinical tests, and thus allows formalization of temporal statements about the progression as well as notions of timed therapies. Certain exemplary models discussed herein can be based on hybrid automata (e.g., with multiple clocks), for which relevant verification and planning algorithms exist.
Owner:NEW YORK UNIV
Construction method and application of caenorhabditis elegans hyperuricemia model and screening method of construction method
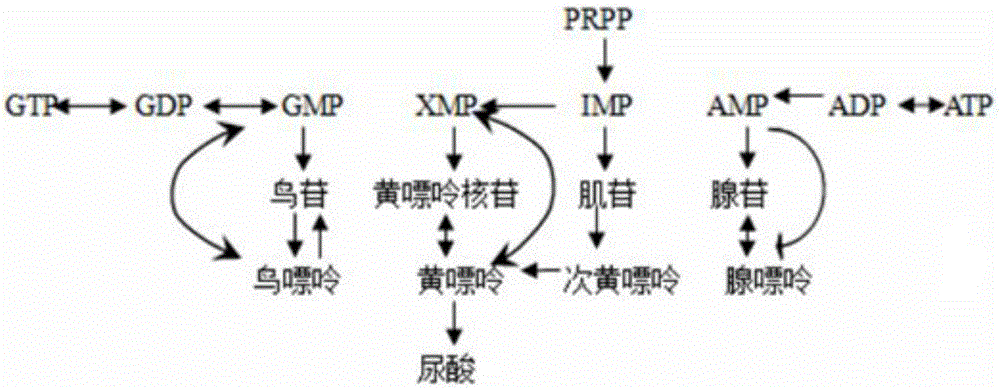
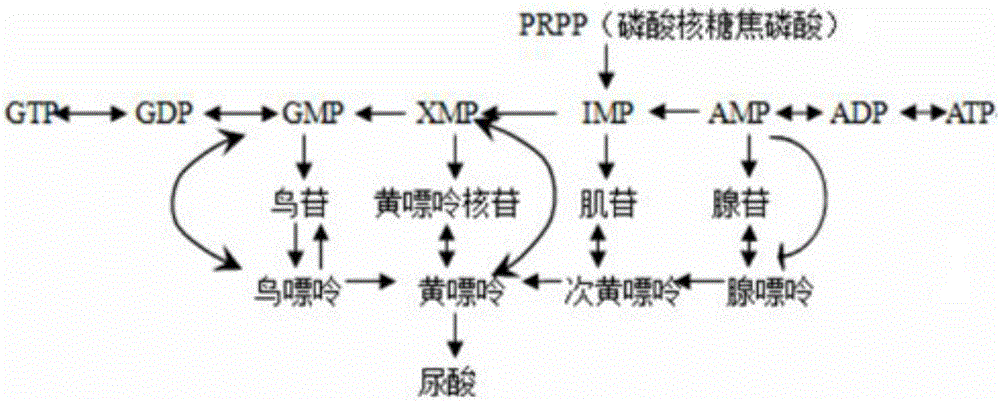
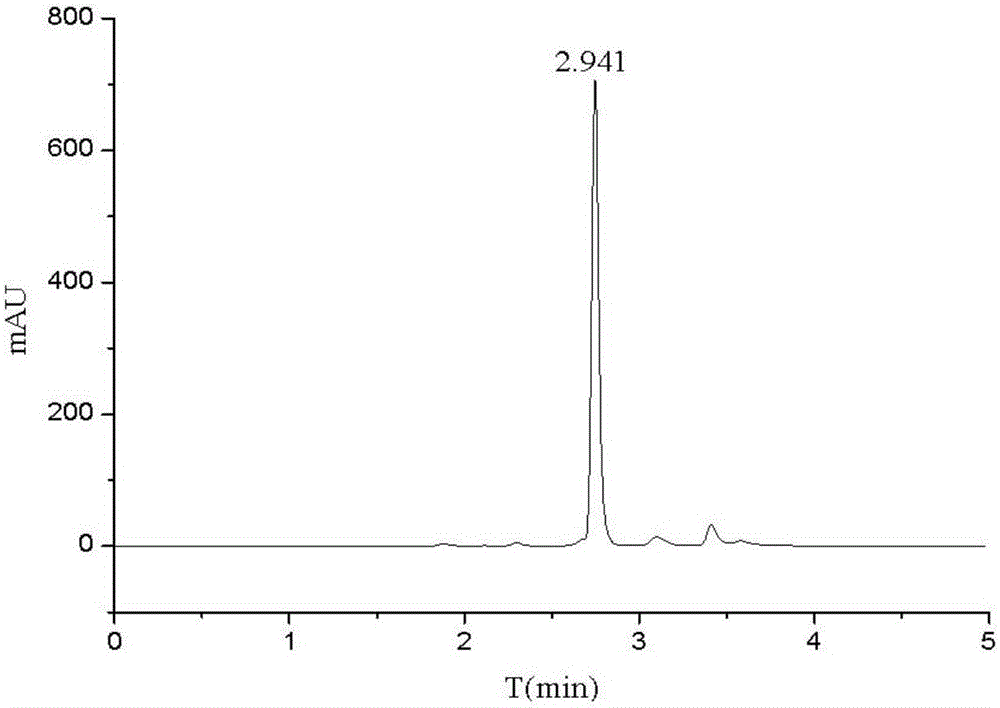
InactiveCN105833295AHigh similarityTypical pathological featuresCompounds screening/testingHeterocyclic compound active ingredientsMedicineMetabolic pathway
The invention relates to a construction method of a caenorhabditis elegans hyperuricemia model. The method comprises the following steps that 1, the uric acid content in caenorhabditis elegans is measured through a high performance liquid chromatography method; 2, an optimal drug delivery and culture mode is determined; 3, an optical drug delivery substance and drug delivery dosage are determined; 4, the optimal drug delivery time of xanthine is determined; 5, series experiment evaluation is conducted on the caenorhabditis elegans hyperuricemia model obtained through the construction method, and the caenorhabditis elegans hyperuricemia model which is low in damage, efficient, stable and reliable is obtained. According to the construction method, the hyperuricemia animal model is constructed by means of the model organism caenorhabditis elegans which has the high conservative property to human genes and has the similar uric acid metabolic pathway with human beings, the model has typical pathological characterization and is stable and reliable, various quantity indexes have the statistical significance, and a powerful tool is provided for large-scale and high-throughput drug screening of gout and the hyperuricemia symptom.
Owner:TIANJIN UNIVERSITY OF SCIENCE AND TECHNOLOGY
2RL chromosome-specific codominant KASP molecular mark for rye and its application
ActiveCN109234449AGood genetic stabilityImproving the efficiency of assisted selection breedingMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceTriticeae
A 2RL chromosome specific codominant KASP molecular mark for rye and its application are disclosed. That KASP molecular mark is SWK2RL1, and the primer of the KASP molecular mark SWK2RL1 comprises anallelic upstream primer of rye, an allelic upstream primer of wheat and a common downstream primer. The nucleotide sequence of the rye allelic upstream primer is shown in SEQ ID NO: 1; the nucleotidesequence of the wheat allelic upstream primer is shown in SEQ ID NO: 2; the nucleotide sequence of the downstream primer is shown in SEQ ID NO: 3. The invention is based on the difference SNP site between rye and wheat, The codominant KASP marker SWK2RL1 of rye universal 2RL chromosome was developed for the first time. This marker has good genetic stability and high resolution, and can be used inwheat chromosome engineering breeding and molecular marker assisted selection breeding. It provides a tool for high throughput detection and tracing of rye chromosome 2RL from different sources in wheat background and accelerates the utilization and transfer of excellent genes on rye chromosome 2RL.
Owner:中国科学院遗传与发育生物学研究所农业资源研究中心
Detection kit and detection method for hepatitis B virus genotyping
InactiveCN102212621AImprove throughputStrong specificityMicrobiological testing/measurementMicroorganism based processesBiotinHepatitis B virus
The invention discloses a detection kit and a detection method for hepatitis B virus genotyping. The kit comprises a primer pair for polymerase chain reaction (PCR) amplification of a hepatitis B virus gene S area and a PreS2 area, a solid phase detection array, and a nucleic acid amplification reaction system and a color development reagent on a solid phase carrier containing DNA polymerase and reaction substrate, wherein a type detection probe and a contrast probe are fixed on the surface of the solid phase detection array. According to the gene chip-based high-flux, high-specificity, quick and easily-popularized detection method using a PCR technology as the principle, the detection probe is fixed on the detection array and hybridized with a PCR amplified gene fragment to be detected; and after the target fragment is identified by the probe on the solid phase carrier, a nucleotide sequence can be synthesized and prolonged, so that the biotin-labeled amplified fragment is fixed on the detection array, is not eluted and further used for color development and display, and the gene type of the hepatitis B virus to be detected can be detected at one time.
Owner:FOURTH MILITARY MEDICAL UNIVERSITY
Separation/purification and pathogenicity identification method for pathogenic bacteria of fusarium root rot of Medicago sativa
InactiveCN106350453AIdentification operation is simpleImprove accuracyFungiMicrobiological testing/measurementPathogenicityCladosporium herbarum
The invention discloses a separation / purification and pathogenicity identification method for pathogenic bacteria of fusarium root rot of Medicago sativa. The method comprises the specific steps of collecting and preparing diseased Medicago sativa plants, carrying out separation and purification on the pathogenic bacteria, carrying out morphological observation on the pathogenic bacteria, carrying out molecular biology identification, constructing an evolutionary tree and carrying out pathogenicity identification. According to the method, the identification on pathogens of root rot of the Medicago sativa is simple in operation and high in accuracy and is simple, convenient and rapid; the separation and purification purity is high, the identification result is reliable, and the strength of pathogenicity of each strain is determined through pathogenicity identification.
Owner:INST OF ANIMAL SCI OF CHINESE ACAD OF AGRI SCI
Primer group and kit for detecting diarrhea-causing parasites through multi-PCR technology
ActiveCN106399486ATo achieve the detection effectGet test results quicklyMicrobiological testing/measurementAgainst vector-borne diseasesDiseaseFood poisoning
The invention provides a primer group for detecting diarrhea-causing parasite genes through the multi-PCR technology. The primer group comprises primers shown as SEQ ID NO.1 and SEQ ID 3-18. The invention further provides a detection method and a kit for detecting the diarrhea-causing parasite genes through the multi-PCR technology. After the technical scheme is adopted, a complete technical means is provided for the rapid and accurate screening of the diarrhea-causing parasite genes, the rapid screening and identification for the diarrhea-causing parasites after the food poisoning event are realized, and a reliable basis is provided for reasonably and properly disposing the diseases.
Owner:北京卓诚惠生生物科技股份有限公司
Primer set and kit for detecting bacillus cereus virulence genes by means of multiplex-PCR and detection method thereof
ActiveCN106434902ATo achieve the detection effectGet test results quicklyMicrobiological testing/measurementMicroorganism based processesFood poisoningPathogenicity
The invention provides a primer set for detecting bacillus cereus virulence genes by means of a multiplex-PCR. The primer set comprises primers shown in SEQ ID NO.1 and 3-24. The invention further provides a detection method and kit for detecting bacillus cereus virulence genes by means of the multiplex-PCR. According to the technical scheme, the perfect technological means is provided for rapid and accurate screening of the bacillus cereus virulence genes, rapid screening and identification of pathogenicity bacillus cereus after a food poisoning event and outbreak of an epidemic situation can be achieved, and the reliable basis is provided for reasonable and appropriate epidemic treatment.
Owner:北京卓诚惠生生物科技股份有限公司
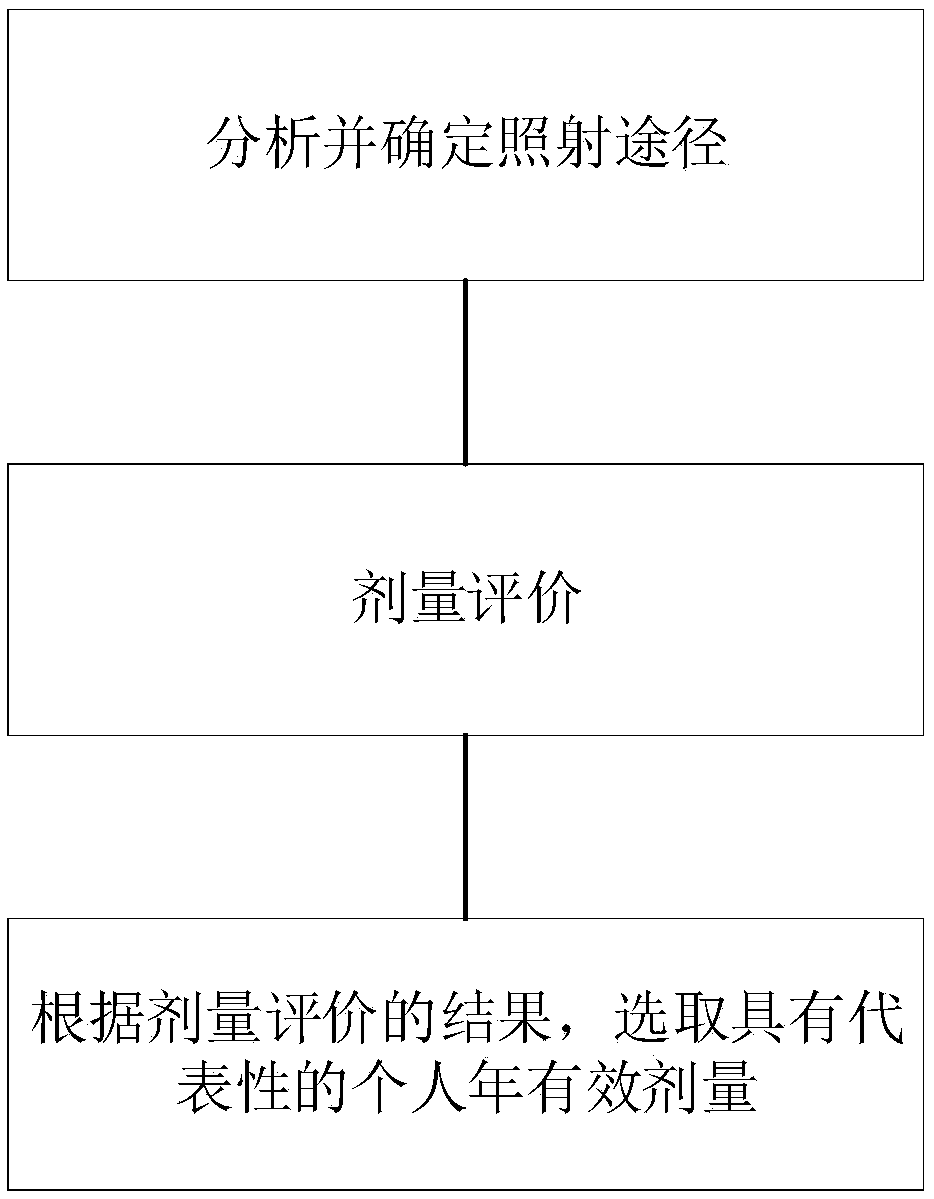
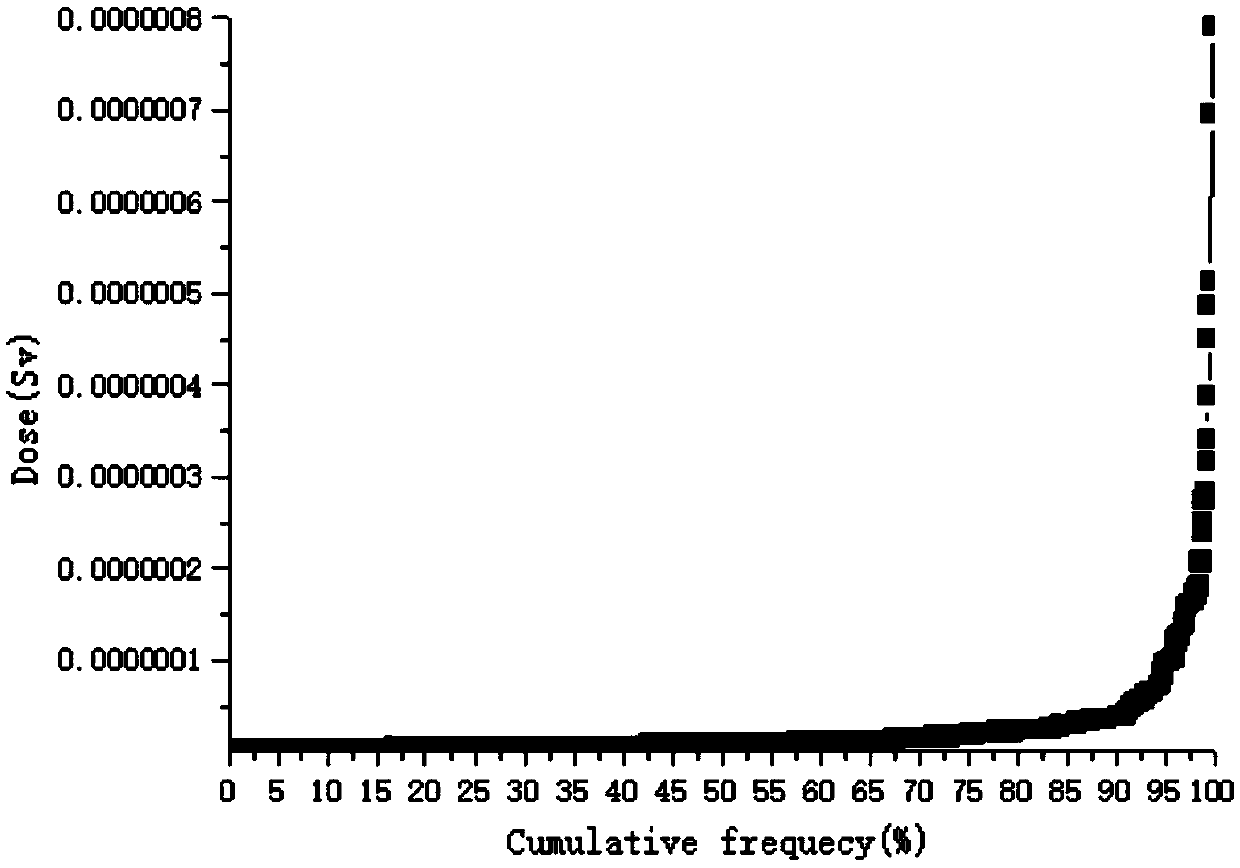
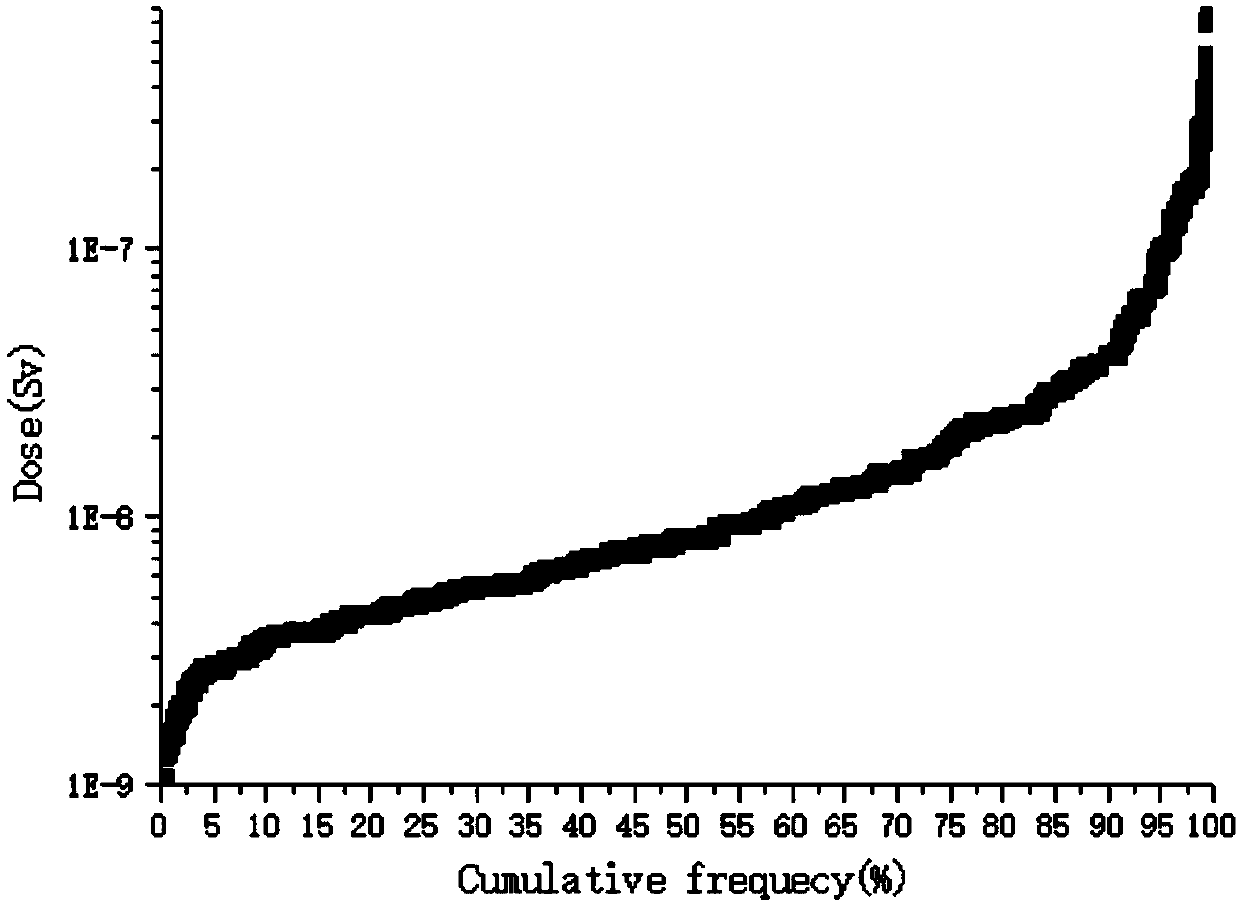
A public dose evaluation method based on a probability theory in radiation environment evaluation
The invention discloses a public dose evaluation method based on a probability theory in radiation environment evaluation. The method comprises the following steps of analyzing and determining an irradiation path, the irradiation path comprising inhaled air internal irradiation, air immersion external irradiation, surface sediment external irradiation, internal irradiation of food-in animal and plant products and dose evaluation, and selecting a representative personal annual effective dose according to the result of the dose evaluation. According to the method, in the public dose estimation process, all the public within the illuminated range are considered, a reasonable personal annual effective dose is selected as a reference dose value, the value serves as an evaluation index in radiation environment evaluation, the conservative property of the evaluation index is further enhanced, and therefore the safety of the public and the environment is guaranteed.
Owner:CHINA INST FOR RADIATION PROTECTION
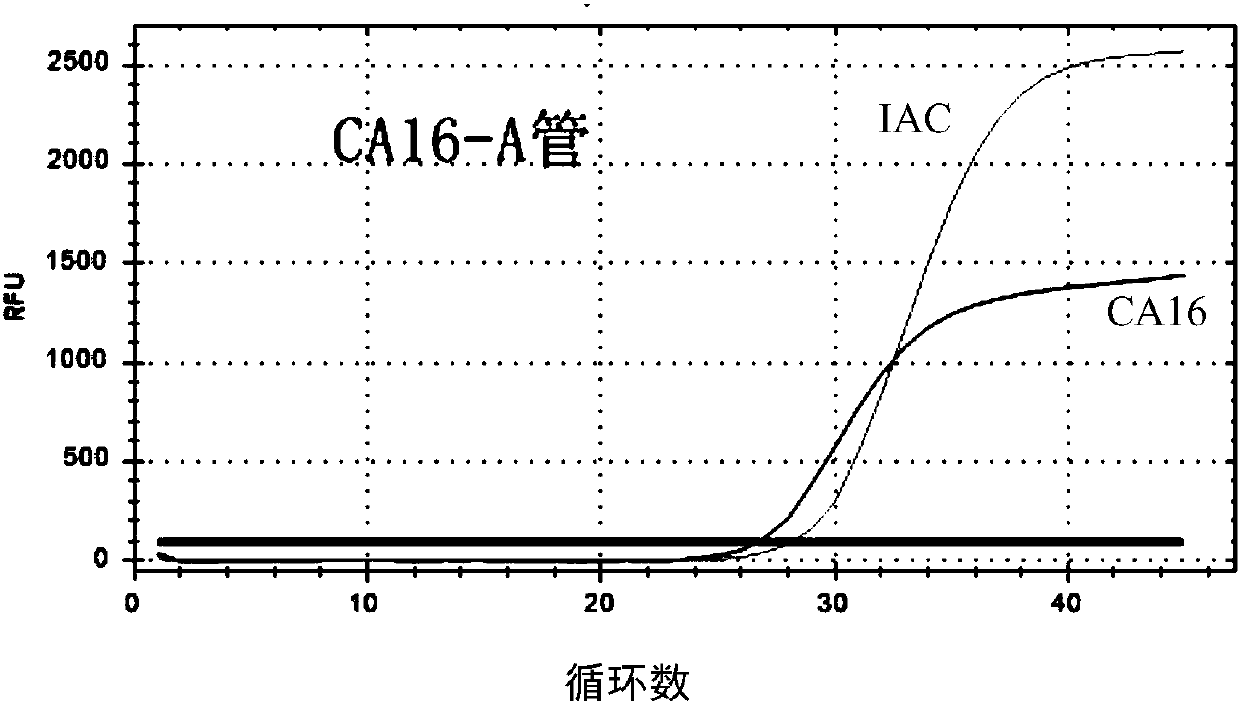
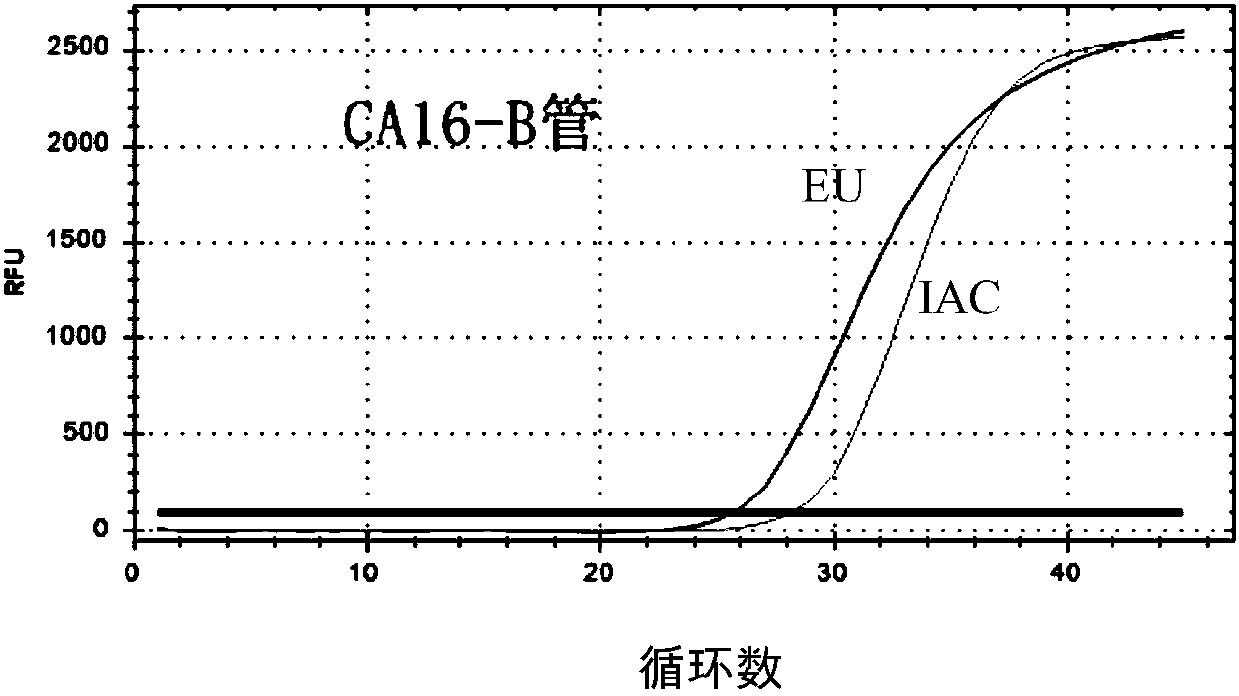
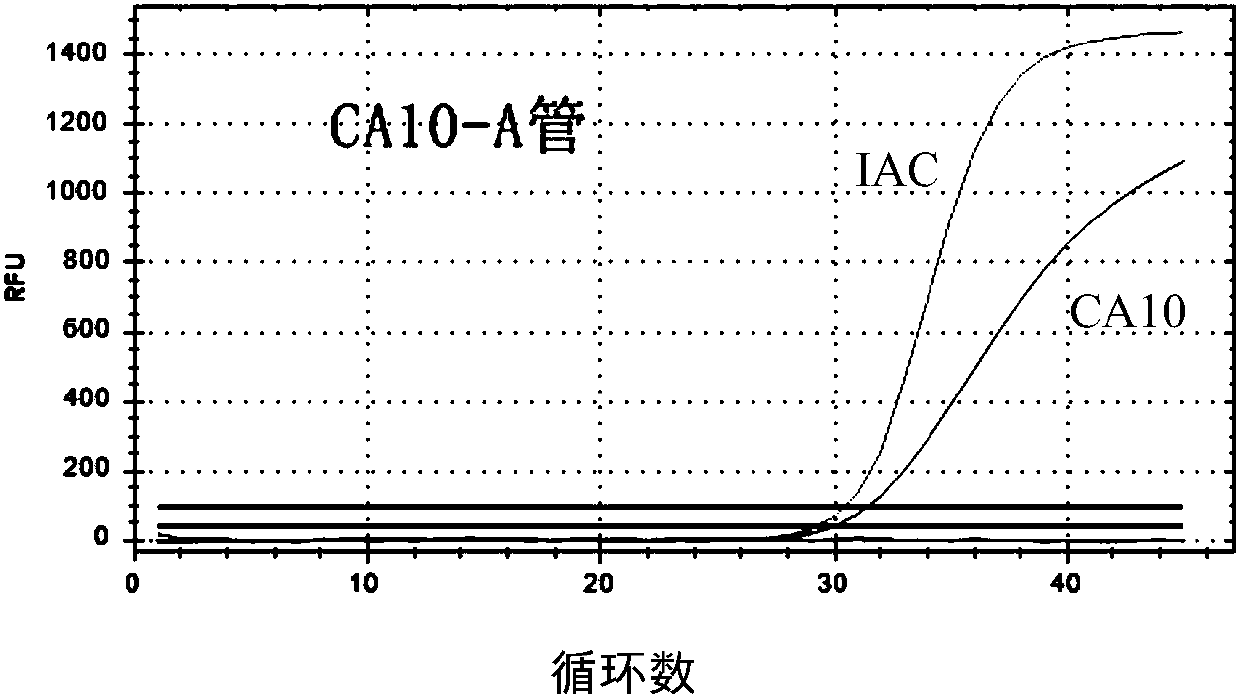
Primer probe set used for detecting coxsackie viruses, enterovirus 71-type and enterovirus universal type through multiple PCR
InactiveCN108034764AEasy to operateRapid implementation of screeningMicrobiological testing/measurementDNA/RNA fragmentationHand-foot-and-mouth diseaseCoxsackie Viruses
The invention relates to a primer probe set used for detecting coxsackie viruses A6 type / A10 type / A16 type / enterovirus 71-type / enterovirus universal type through multiple PCR. The invention also provides a kit used for detecting coxsackie viruses A6 type / A10 type / A16 type / enterovirus 71-type / enterovirus universal type through multiple quantitative fluorescence PCR. The kit comprises the primer probe set. The above technical scheme provides a complete solution scheme for rapidly screening coxsackie viruses A6 type / A10 type / A16 type / enterovirus 71-type / enterovirus universal type in food, can increase emergent processing for hand-foot-and-mouth disease and comprehensive control capability, provides the necessary technical guarantee for reducing large-scope spreading of hand-foot-and-mouth disease in population, and eliminates the social influence and economic loss due to outbreak of infectious diseases.
Owner:北京卓诚惠生生物科技股份有限公司
Avian leukosis P27 monoclonal antibody hybridoma cell strain and application thereof
InactiveCN103243077AHighly conservativeIncreased sensitivityImmunoglobulins against animals/humansImmunoglobulins against virusesSerodiagnosesLeucosis
The invention belongs to the technical field of animal epidemic disease serological diagnosis and relates to an avian leukosis P27 monoclonal antibody hybridoma cell strain and application thereof. The avian leukosis P27 monoclonal antibody obtained by secreting the hybridoma cell strain can be applied to detection of the avian leukosis P27, the 96-pore elisa plate is coated by the P27 specific polyclonal antibody, the P27 monoclonal antibody is labeled by alkaline phosphatase, and according to the avian leukosis P27 monoclonal antibody and the P27 monoclonal antibody labeled by alkaline phosphatase, the detection sensitivity, specificity and stability are effectively improved. A high-efficiency and sensitive detection mode is provided for a novel variety which is used for selecting, purifying and breeding avian leukosis positive chickens and has high genetic resistance, and the avian leukosis P27 monoclonal antibody hybridoma cell strain is low in cost, easy and convenient to operate and is suitable for popularization and application of animal husbandry production.
Owner:LANZHOU INST OF ANIMAL SCI & VETERINARY PHARMA OF CAAS +2
Fluorescence quantitative PCR (polymerase chain reaction) detection method of duck hepatitis B virus, and reagent
InactiveCN103114153ANot easy to mutateAvoid Vulnerable DefectsMicrobiological testing/measurementFluorescence/phosphorescenceFluorescenceHepatitis B virus
The invention discloses a fluorescence quantitative PCR (polymerase chain reaction) detection method of duck hepatitis B virus (DHBV), and a reagent. Nucleic acid sequences in a conservative region most consistent to a DHBV Core region are used as a template to design and synthesize a primer and a fluorescent probe. A pGEM-T / DHBV Core recombinant plasmid is established to prepare a PCR standard product. A reaction system and amplification conditions are optimized. The fluorescence quantitative PCR detection method provided by the invention has the advantages of strong versatility, high sensitivity, strong specificity, good reproducibility and the like, and is suitable for detection of DHBV DNA (deoxyribonucleic acid) of different duck species in different regions.
Owner:THE FIRST AFFILIATED HOSPITAL OF MEDICAL COLLEGE OF XIAN JIAOTONG UNIV
Epitope polypeptide combination capable of inducing immunity and application thereof
PendingCN113372417AVerify immune response propertiesHighly conservativeSsRNA viruses positive-senseViral antigen ingredientsAntigen epitopeCtl epitope
The invention discloses an epitope polypeptide combination capable of inducing immunity and application thereof, belongs to the technical field of biology, and aims to carry out molecular design of related vaccines by utilizing immunoinformatics on the basis of epitope analysis optimization. Based on a structural antigen epitope vaccine design strategy, a B cell epitope, a Th epitope and a CTL epitope on a new coronavirus S protein are determined through immunoinformatics to induce a main neutralizing antibody, activate cellular immune response and induce body fluid and cellular immune balance. The epitope vaccine is designed through connection of a molecular adjuvant and candidate antigen epitopes, antigenicity, physicochemical properties, protein secondary structure and tertiary structure modeling of the epitope vaccine are analyzed, vaccine conformation B cell epitopes are analyzed by means of a structural biological tool, and immune response characteristics of the vaccine are verified through molecular docking with TLR4 and immune response simulation stimulation. Information analysis results show that the designed candidate epitope combination has well balanced humoral immune and cellular immune response capabilities.
Owner:SHANTOU UNIV MEDICAL COLLEGE
Polypeptide, immunogenicity conjugate and influenza vaccine
InactiveCN107488217AReduce morbidityAvoid deathSsRNA viruses negative-senseAntibody mimetics/scaffoldsVirus influenzaImmunogenicity
The invention discloses a polypeptide, an immunogenicity conjugate and an influenza vaccine, and relates to the bio-technical field. The polypeptide is any one of a), b) or c): a) a polypeptide with the amino acid sequence as shown in SEQ ID NO.1; b) fused protein obtained by connecting a label to the end N and / or the end C of the polypeptide with the amino acid sequence as shown in SEQ ID NO.1; c) a polypeptide which is obtained by substitution and / or deletion and / or adding of one or several amino acid residues for the amino acid sequence as shown in SEQ ID NO.1 and has the same function. The polypeptide can be used for preventing or treating influenza virus, and can be used as an active component for preparing a medicine for preventing or treating the influenza virus. The immunogenicity conjugate disclosed by the invention can improve the immunogenicity and the immune effect on an antigen at a conformational dependency site.
Owner:华兰生物疫苗股份有限公司
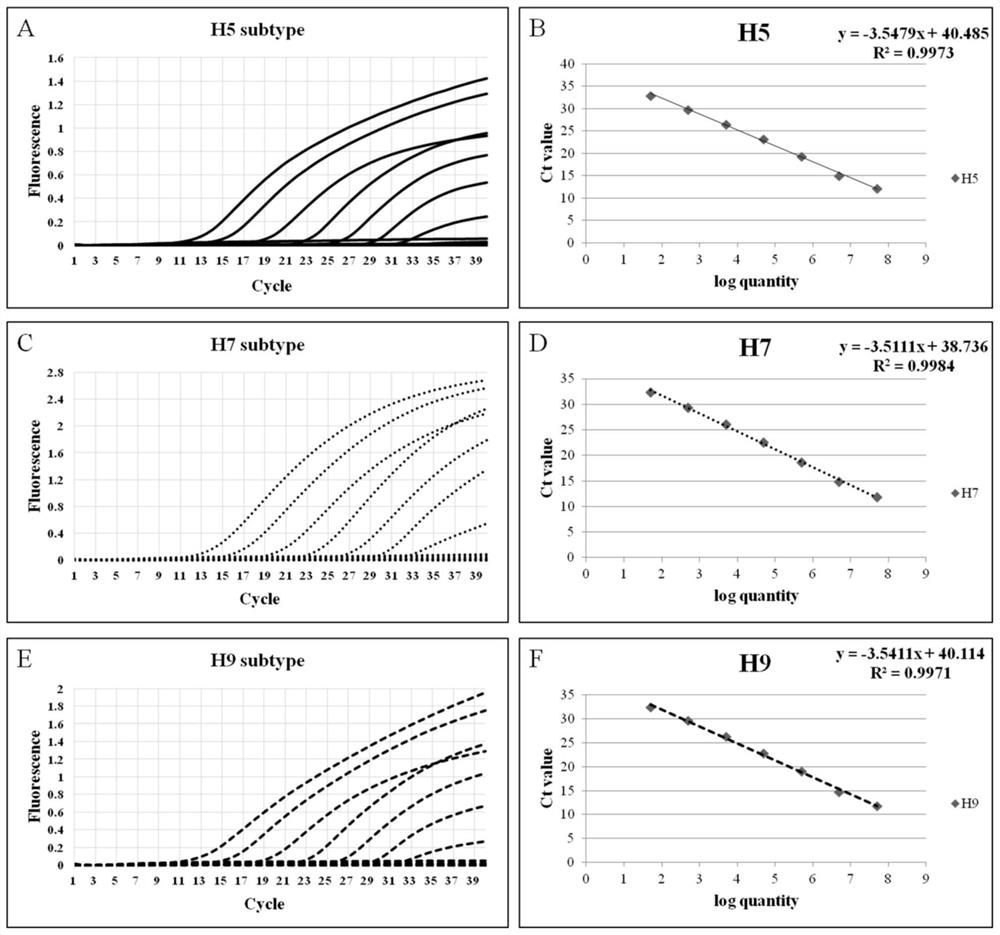
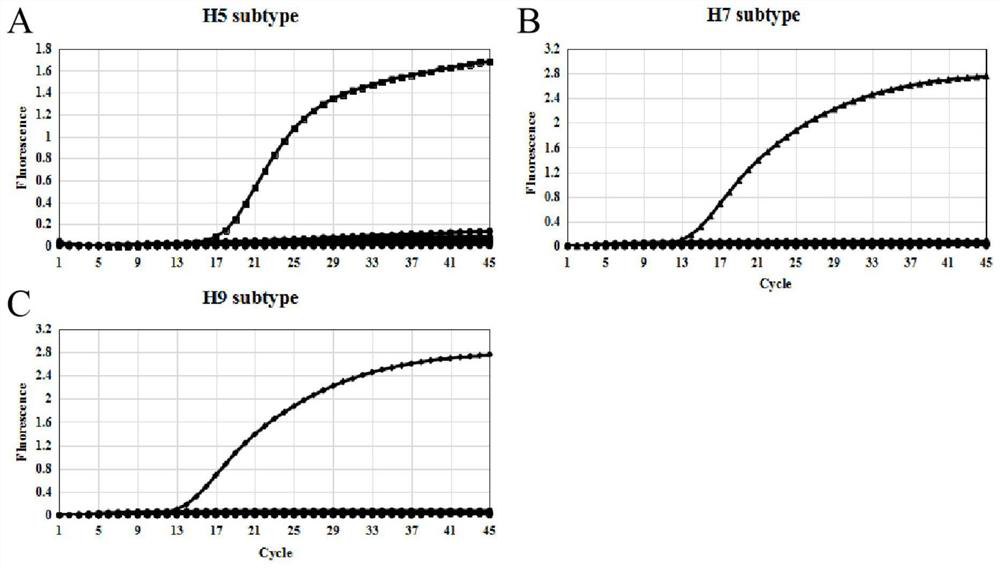
Avian influenza virus H5, H7 and H9 subtype triple fluorescent quantitative RT-PCR detection kit and application thereof
PendingCN112176112AGreat application potentialImprove detection efficiencyMicrobiological testing/measurementDNA/RNA fragmentationMedicineViral test
The invention discloses an avian influenza virus H5, H7 and H9 subtype triple fluorescent quantitative RT-PCR detection kit, and provides an optimal reaction system and reaction program of the kit anda use method thereof. The specificity of the kit reaches 100%, and the kit can specifically detect H5, H7 or H9 subtype AIV in the detection process of H1-H13 subtype AIV, NDV, IBV, IBDV and other common avian viruses. The detection sensitivity of the kit on the H5, H7 and H9 subtype avian influenza viruses in a sample can reach 50 copies / mu L, 50 copies / mu L and 50 copies / mu L. On the premise ofensuring single-tube multi-detection high detection efficiency and low cost, the kit has the characteristics of low cross contamination rate, simplicity and convenience in operation, fluorescence monitoring property, quickness, accuracy, high specificity, high sensitivity and high stability.
Owner:NANJING AGRICULTURAL UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com