Novel coronavirus antigen epitope and application thereof
A technology of antigenic epitopes and coronaviruses, applied in the direction of viral antigen components, viruses, viral peptides, etc., can solve problems such as affecting the immune response of viruses and changing the immunogenicity of viruses
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Example 1 Antigen epitope calculation
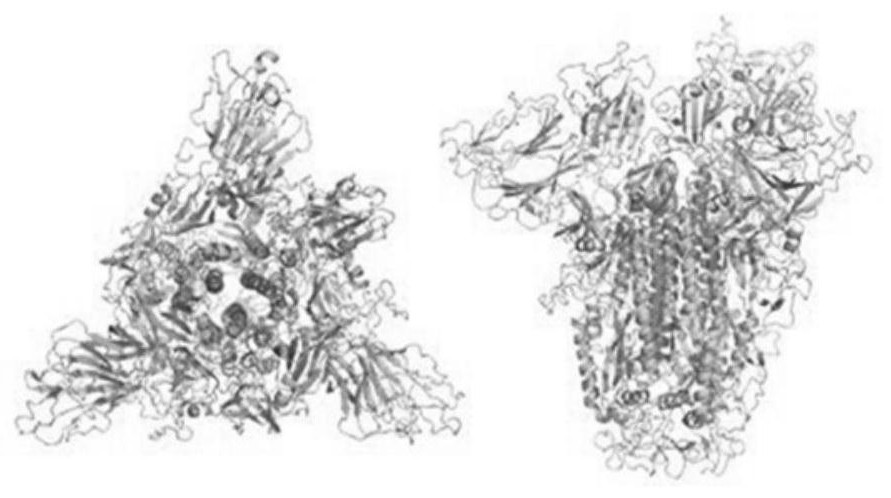
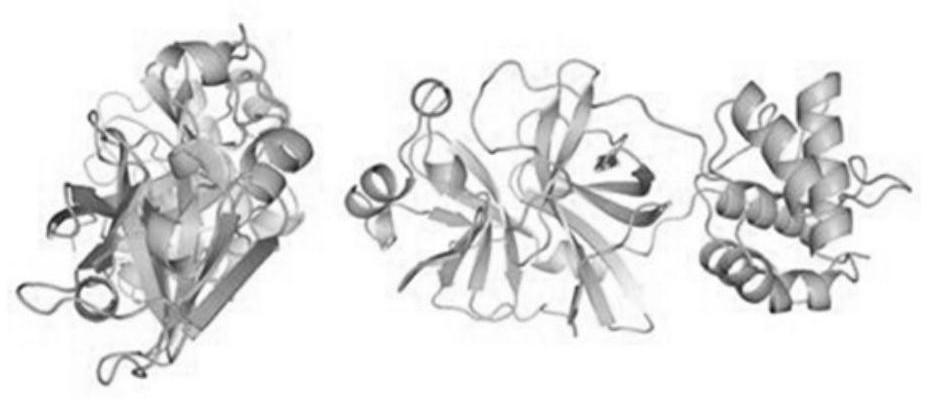
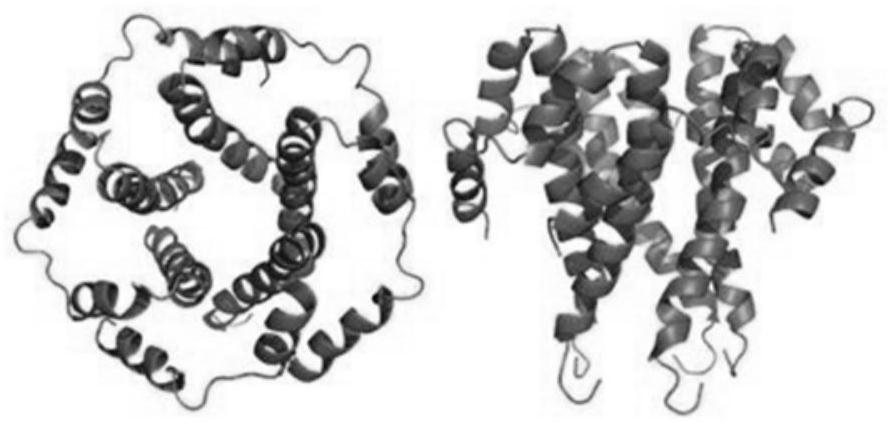
[0075] This embodiment is based on the whole genome nucleic acid sequence of SARS-CoV-2 (GeneBank: MN988669.1), and the original pdb files of S protein, M protein, E protein and N protein are established by the method of homology modeling; in the original pdb file Based on adding water and adjusting the pH of Cl - and Na + , run the molecular dynamics simulation program to obtain the pdb file under the state of human body temperature (310K); based on the pdb file under the state of human body temperature, the molecular dynamics software Gromacs is used to calculate the all-atom structure of the protein, and the following is obtained: Figure 1A , Figure 1B , Figure 1C with Figure 1D The molecular orbital energy level and spatial structure information of the S protein, M protein, E protein and N protein shown;
[0076] Based on the spatial structure information of the structural models of S protein, M protein, E protein and ...
Embodiment 2
[0077] Homology Analysis of Example 2 Epitope
[0078] The amino acid sequences of the S protein, M protein, E protein, and N protein of SARS-CoV-2 were compared with the corresponding amino acid sequences of the S protein, M protein, E protein, and N protein of SARS-CoV and RaTG13 using the ClustalW program to gradually compare To compare the methods, the steps are as follows:
[0079] (1) Input the protein sequence, set the Gap Open Penalty parameter to 10, the Gap Extension Penalty parameter to 0.05, and the scoring matrix to BLOSUM, and compare the three sequences pairwise to construct a relationship matrix;
[0080] (2) Generate a phylogenetic tree based on the distance matrix, start from the two closest sequences, gradually introduce adjacent sequences to construct an alignment, and obtain a protein homology analysis diagram;
[0081] (3) Mark the selected antigenic epitope on the protein homology analysis map, and analyze the conservation of the antigenic epitope among...
Embodiment 3
[0086] Example 3 Utilizes HBc-S to prepare a VLP vaccine displaying antigenic epitopes
[0087] Synthesize the epitope-SpyTag, mix it with the HBc VLP expressing SpyCatcher at a molar ratio of 3:1, add 3 times the volume of citrate buffer, and place it at room temperature for 1-3h; then the mixture is treated with 100kDa Cut-off membrane ultrafiltration to remove unreacted polypeptides (citric acid reaction buffer as ultrafiltration liquid) to obtain HBc VLPs vaccines displaying each antigenic epitope polypeptide.
[0088] The result is as Figure 3A As shown, the size of the HBc-S monomer is about 27kDa, and after binding to the polypeptide, HBc-S-P is slightly larger than the HBc-S monomer, indicating that each polypeptide is bound to the surface of the HBc-S carrier protein; all the strips of HBc-S-P The bands all moved up, indicating that the binding efficiency of HBc-S to the polypeptide is extremely high.
[0089] Subsequently, the assembly of HBc-S-P was detected by t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com