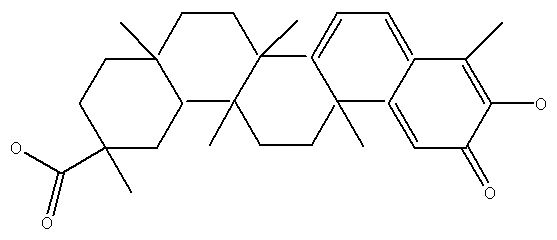
Application of tripterine in preparation of anti-coronavirus products
A technology of tripterygne and coronavirus, which is applied in the field of medicine and can solve problems such as no anti-virus research reports of tripterygium
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1 Molecular docking with the new coronavirus S protein as the target protein
[0022] The PDB database downloads the new coronavirus Spike protein in PDB format, adding hydrogen atoms and charges to the tertiary structure model of the target protein S protein, and repairing missing amino acid residues. The AMBER FF99 force field in the SYBYL X-1.2 software package was selected for energy optimization. First, select the AMBER FF99 force field to perform 1000 iterations of optimization using the steepest descent (SD) method. Then, the conjugated gradient method (conjugated gradient, CG for short) was used to optimize to a convergent gradient of 0.05 kcal / (Å mol). The 2D structure of tripterine was drawn by Chem3D, the 3D structure of tripterine was generated using Chem3D software, and the energy was minimized, and all the small molecule ligands used were saved in mol2 format. Check the molecular structure of the ligand, convert the molecular planar structure int...
Embodiment 2
[0023] Example 2 Molecular docking with ACE2 as the target protein
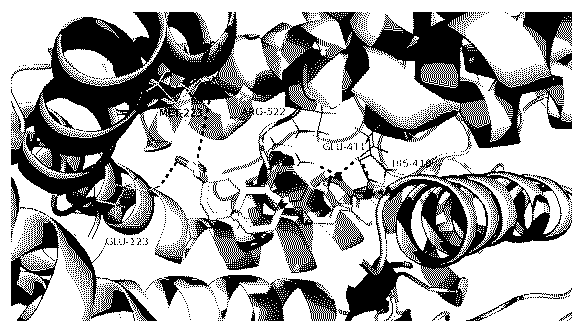
[0024] PDB database download The ACE2 structure is in PDB format, adding hydrogen atoms and charges to the tertiary structure model of the target protein ACE2, and repairing missing amino acid residues. The AMBER FF99 force field in the SYBYL X-1.2 software package was selected for energy optimization. After iterative optimization, the conjugate gradient method was used to optimize to a convergent gradient of 0.05 kcal / (Å mol). Draw the structure of tripterine, perform energy minimization, and save all the small molecule ligands used in mol2 format. Then add hydrogen, add charge, and optimize energy. The interaction force between the ligand molecule and the protein is calculated based on the Total score. The docking results showed that the total score of tripterine and ACE2 was 5.249 (attached image 3 ), tripterine has obvious combination with ACE2.
Embodiment 3
[0025] Example 3 Molecular docking with novel coronavirus RNA-dependent RNA polymerase as target protein
[0026] The PDB database downloads the structure of the new coronavirus RNA-dependent RNA polymerase in PDB format, adding hydrogen atoms and charges to the tertiary structure model of the target protein RNA-dependent RNA polymerase, and repairing the missing amino acid residues. After energy optimization, iterative optimization was performed, and then CG was used to optimize to a convergence gradient of 0.05 kcal / (Å mol). Draw the structure of tripterine, perform energy minimization, and save all the small molecule ligands used in mol2 format. Check the molecular structure of the ligand, add hydrogen to the molecular structure, add charges, and optimize the energy. The interaction force between the ligand molecule and the protein is calculated based on the Total score. The docking results showed that the total score of tripterine and novel coronavirus RNA-dependent RNA ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com