Regioselective bacterium nitroreductase gene and application thereof
A selective and reductase technology, applied in the field of enzyme engineering, can solve problems such as poor catalytic effect and lack of regioselectivity in reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 Gene acquisition and expression vector construction of mutant nitroreductase F124W, N71S / F124W, F123A / F124W and N71S / F123A / F124W
[0025] 1. Cloning of wild-type nfsB gene and construction of cloning vector
[0026] According to the nfsB gene sequence in the E.coli K12 strain (Invitrogen) provided by NCBI, primers (nfsB-WT-F: 5'-GGAATTC CATATG GATATCATTTCTGTCGCCTTAAAG-3',nfsB-WT-R:5'-G GAATTCTTACACTTCGGTTAAGGTGATGTT-3'), where the underlined part represents the gene sequence corresponding to the restriction site. Using the extracted E.coli K12 genomic DNA as a template, PCR amplification was performed. Take 10 μl of the PCR reaction product and run agarose gel electrophoresis to verify that the electrophoresis band is about 650bp in size consistent with the nfsB gene. Use TaKaRa Agarose Gel DNA Purification Kit Ver.3.0 to cut the gel to recover the above electrophoresis target band, then digest the recovered product with EcoR I / Nde I double digestion, use...
Embodiment 2
[0043] Example 2. Expression and purification of bacterial nitroreductase mutants
[0044] 1. Induced expression of nitroreductase mutants
[0045] The recombinant plasmids pET28a-nfsB-F124W, pET28a-nfsB-N71S / F124W, pET28a-nfsB-F123A / F124W and pET28a-nfsB-N71S / F123A / F124W were transformed into E. coli E. coli BL21 (DE3) competent Cells (Invitrogen), plated and incubated overnight at 37°C. Pick positive clones and name them as expression strains E.coli BL21(DE3)-nfsB-F124W, E.coli BL21(DE3)-nfsB-N71S / F124W, E.coli BL21(DE3)-nfsB-F123A / F124W and E.coli BL21(DE3)-nfsB-N71S / F123A / F124W were inoculated in LB liquid medium (containing 50 μg / ml Kana) and shaken overnight at 37°C. Transfer the seed culture solution to 100ml LB expansion medium (containing 50μg / ml Kana) at 1% inoculum size, and culture it on a shaker at 37°C until OD 600 at 0.4-0.6. Add final concentration of 0.5mM IPTG to induce expression at 30°C for 6h. After the cultivation, the fermentation broth was centrifu...
Embodiment 3
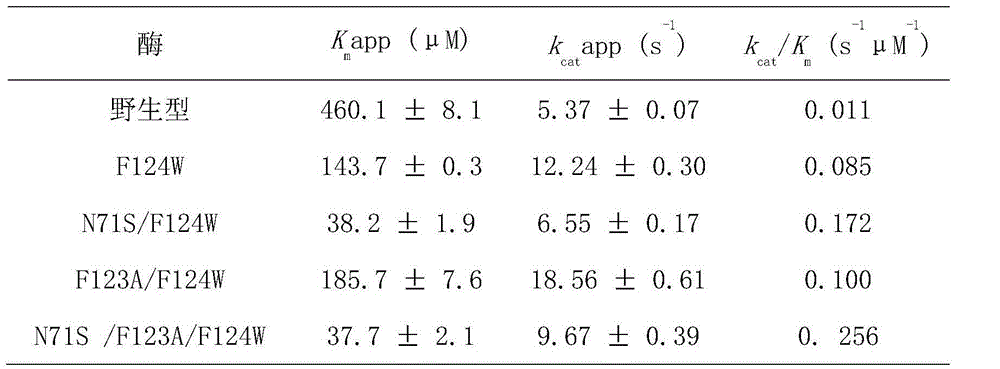
[0059] Example 3. Activity determination of bacterial nitroreductase mutants
[0060] The recombinant protein eluted with 250mM imidazole was mixed according to the ratio of molar concentration ratio protein:FMN=1:10, and placed in a 4°C refrigerator for incubation for 2 hours. After incubation, the recombinant protein was desalted into 20 mM Tris-HCl buffer (pH 7.0) for subsequent experiments such as protein activity detection.
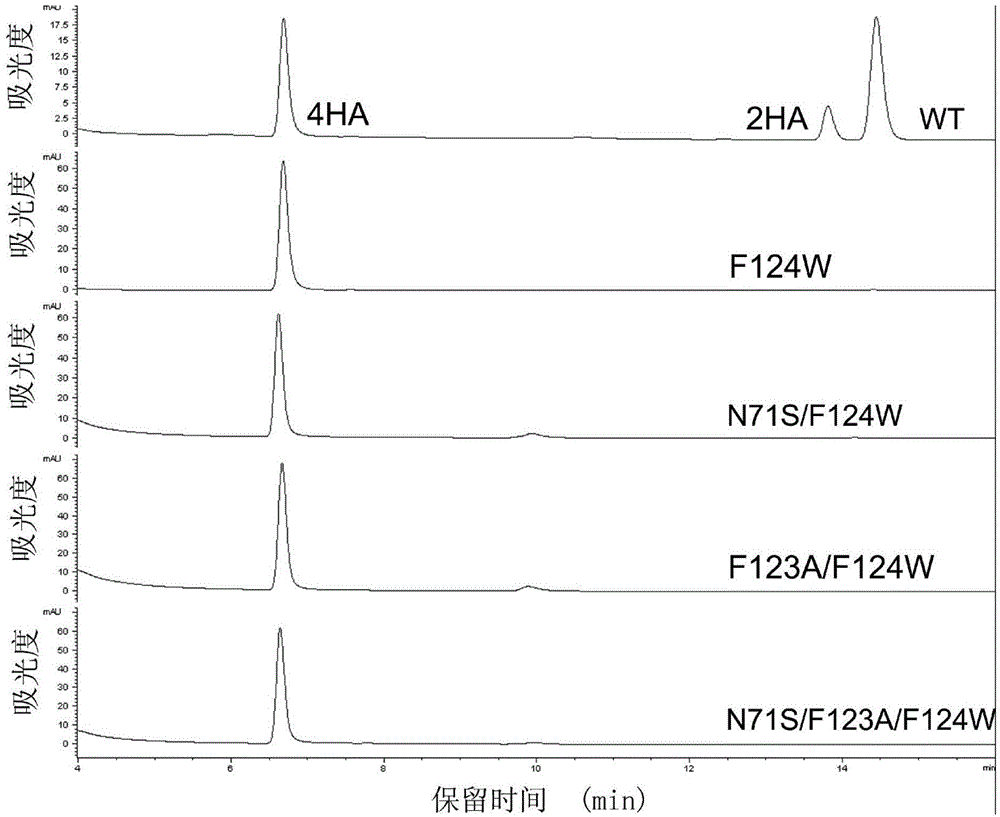
[0061] 1. HPLC analysis of the mutant's regioselectivity for CB1954 reduction
[0062] The regioselectivity of mutants NfsB-F124W, NfsB-N71S / F124W, NfsB-F123A / F124W and NfsB-N71S / F123A / F124W for the reduction of substrate CB1954 was analyzed by high performance liquid chromatography (HPLC) Agilent 1200. The HPLC enzyme-catalyzed reaction system contains: 0.1mM CB1954, 0.2mM NADH, 200nM mutant enzyme, the reaction buffer is 20mM Tris-HCl pH7.0, and react at room temperature for 5-15 minutes. Since CB1954 and its reaction product were unstable when h...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com