Virtual screening method of targeted IKK beta drug
A virtual screening and drug technology, applied in bioinformatics, instruments, and used to analyze two-dimensional or three-dimensional molecular structures, etc. The effect of application value and prospect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
experiment example 1
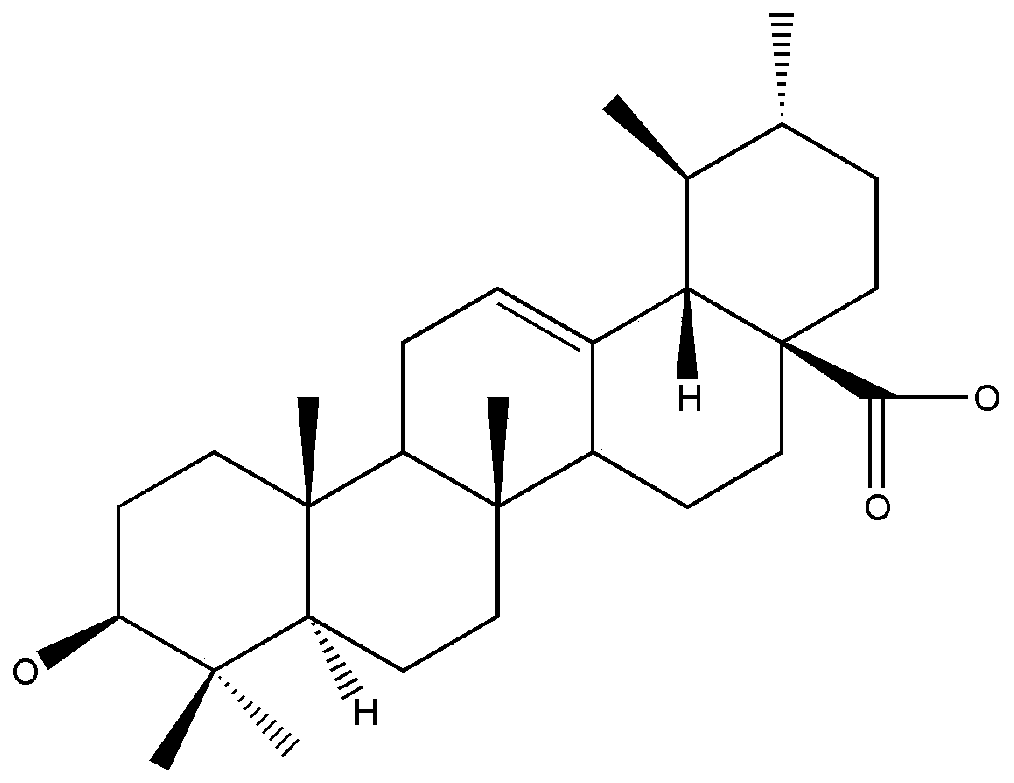
[0035] Analysis of the binding effect of ursolic acid and IKKβ by iGEMDOCK molecular docking software
[0036] IKKβ is a receptor, and its three-dimensional crystal structure is derived from the PDB protein database (ID: 4KIK); ursolic acid is a ligand. The iGEMDOCK molecular docking software directly imports receptors and ligands, docking parameter settings: Population Size is set to 200, Generations is set to 70, Number of solutions is set to 2 (below same). The binding energy between ursolic acid and IKKβ was found to be -98.9kcaj / mol, indicating that ursolic acid and IKKβ have a certain binding ability.
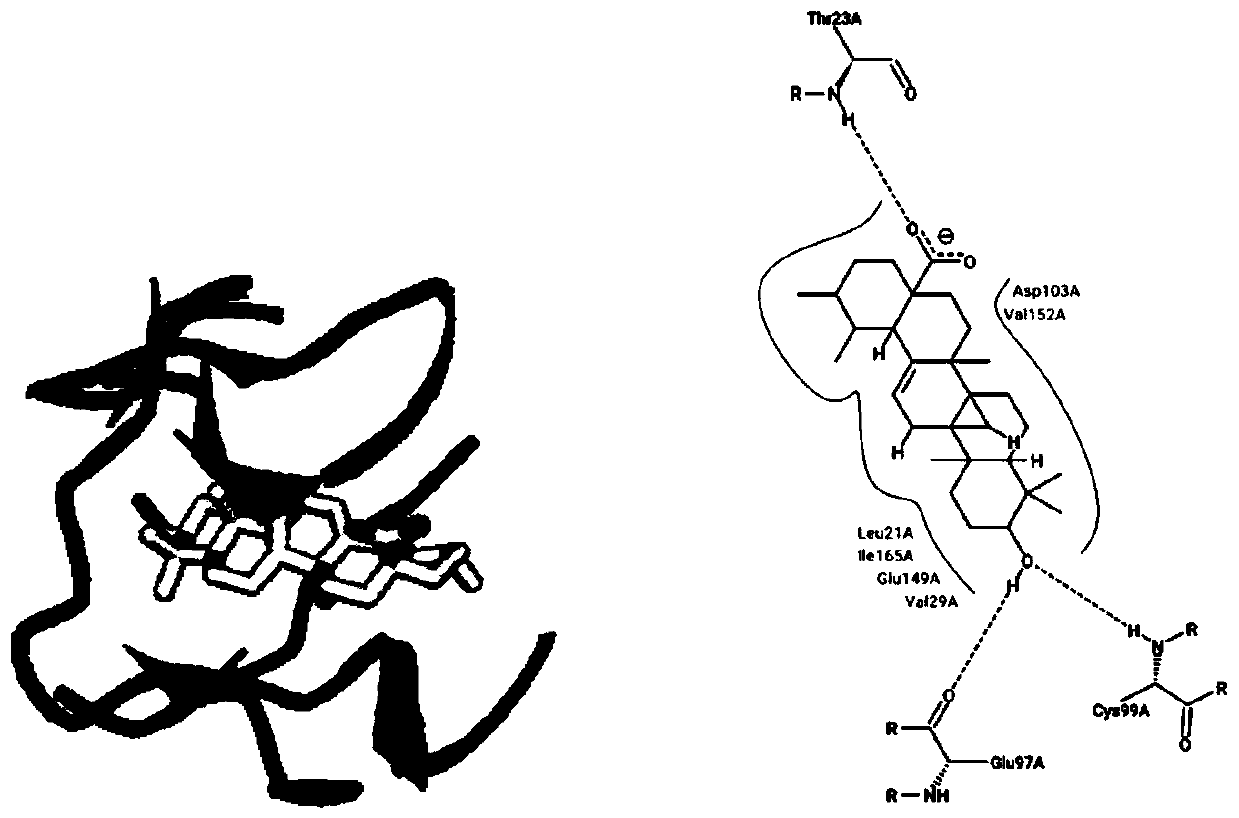
[0037] image 3 It is a schematic diagram of molecular docking analysis between ursolic acid and IKKβ. The binding sites between ursolic acid and IKKβ are Leu21, Thr23, Glu97, and Cys99, which are located in the active pocket of IKKβ. , Cys99 site produces hydrogen bonding, and the carboxyl group on the C28 position of ursolic acid produces hydrogen bonding with the Le...
Embodiment 2
[0039] The iGEMDOCK molecular docking software analyzes the binding effect of different side chain groups on the C3 position of ursolic acid and IKKβ
[0040] According to the results of Example 1, the hydroxyl group on the C3 position of ursolic acid is modified with a side chain group, and the selected side chain group R1~11 is shown in Figure 4 (The hydrogen in the side chain group has been omitted, the same below). The iGEMDOCK molecular docking software was used to analyze the binding effect of the modified ursolic acid and IKKβ respectively.
[0041] Taking the side chain groups R2, R6, R9 as an example, Figure 5-7 Schematic diagrams of molecular docking analysis of modified ursolic acid and IKKβ, respectively. It can be seen from the figure that R2, R6, and R9 can all form hydrogen bonds with Glu97 and Cys99. Among them, the amino hydrogen atom of R2 penetrates into the ATP binding region and forms a hydrogen bond with Glu97 and Cys99; the carboxyl hydrogen atom of R...
Embodiment 3
[0047] The iGEMDOCK molecular docking software analyzes the binding effect of different side chain groups on the C28 position of ursolic acid and IKKβ
[0048] According to the result of embodiment 1, the carboxyl group on the C28 position of ursolic acid is carried out side chain group transformation, the side chain group R1 '-14' of selection sees Figure 8 , using iGEMDOCK molecular docking software to analyze the binding effect of the modified ursolic acid and IKKβ respectively.
[0049] Take the side chain group R1', R2', R14' as an example, Figures 9 to 11 Schematic diagrams of molecular docking analysis of modified ursolic acid and IKKβ, respectively. It can be seen from the figure that both the ether group oxygen of R1' and the ester group oxygen of R2' form hydrogen bonds with Thr23, indicating that the side chain group at the C28 position can go deep into the Gly ring and does not interact with the ATP binding region. The carbonyl oxygen atom and amino hydrogen atom...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com