Primer set of wheat leaf rust est-ssr molecular marker and its detection method and application
A technology of wheat leaf rust and molecular markers, applied in biochemical equipment and methods, microbial measurement/testing, recombinant DNA technology, etc., can solve the problems of strong randomness, difficult conditions to control, cumbersome steps, etc., and achieve simple operation , convenient detection, rich polymorphism effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Obtaining of RNA-seq data of wheat leaf rust and EST-SSR primer synthesis
[0042] The RNA of wheat leaf rust fungus was extracted, and the samples were submitted to Huada (Shenzhen) Company for RNA-Seq sequencing.
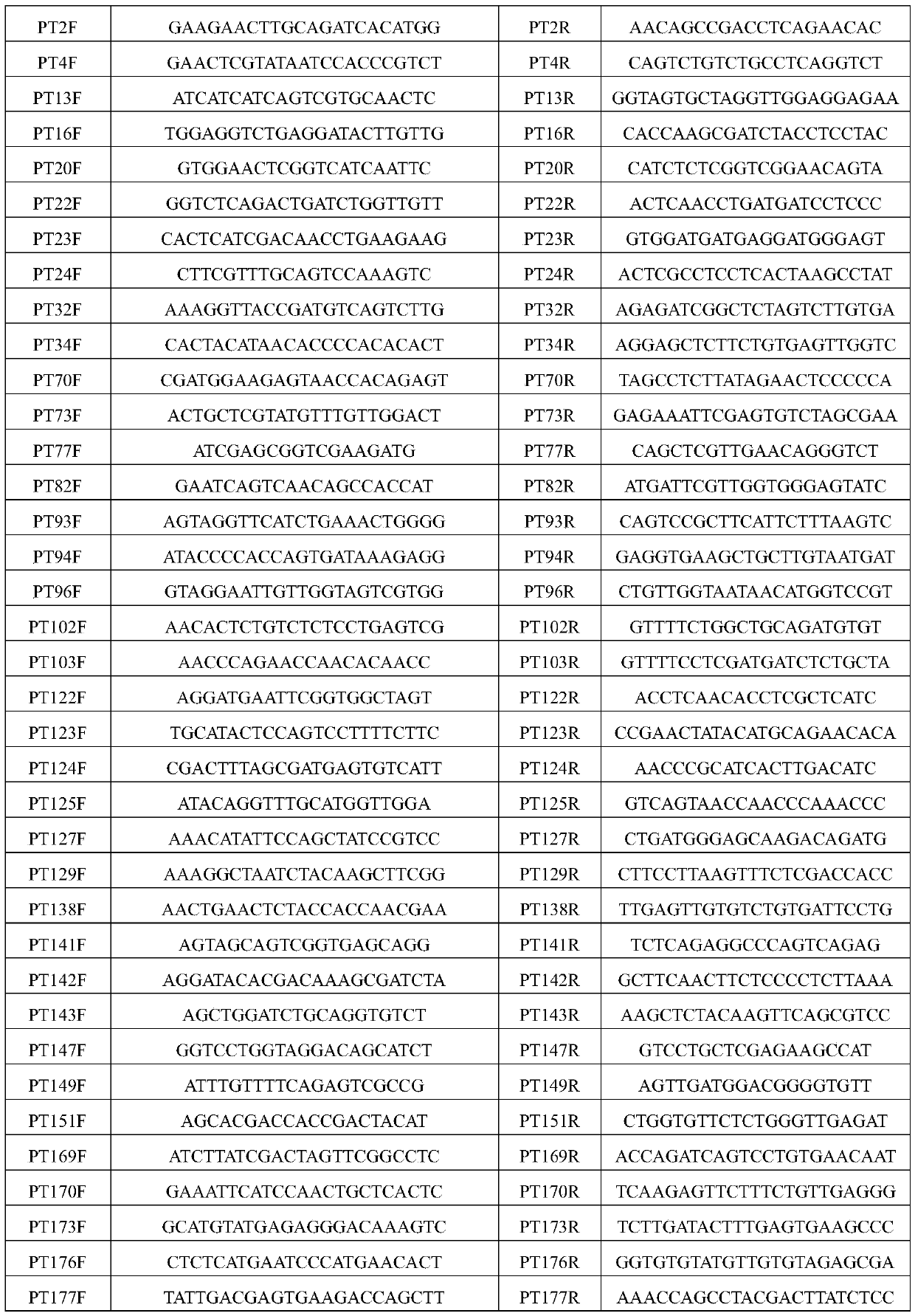
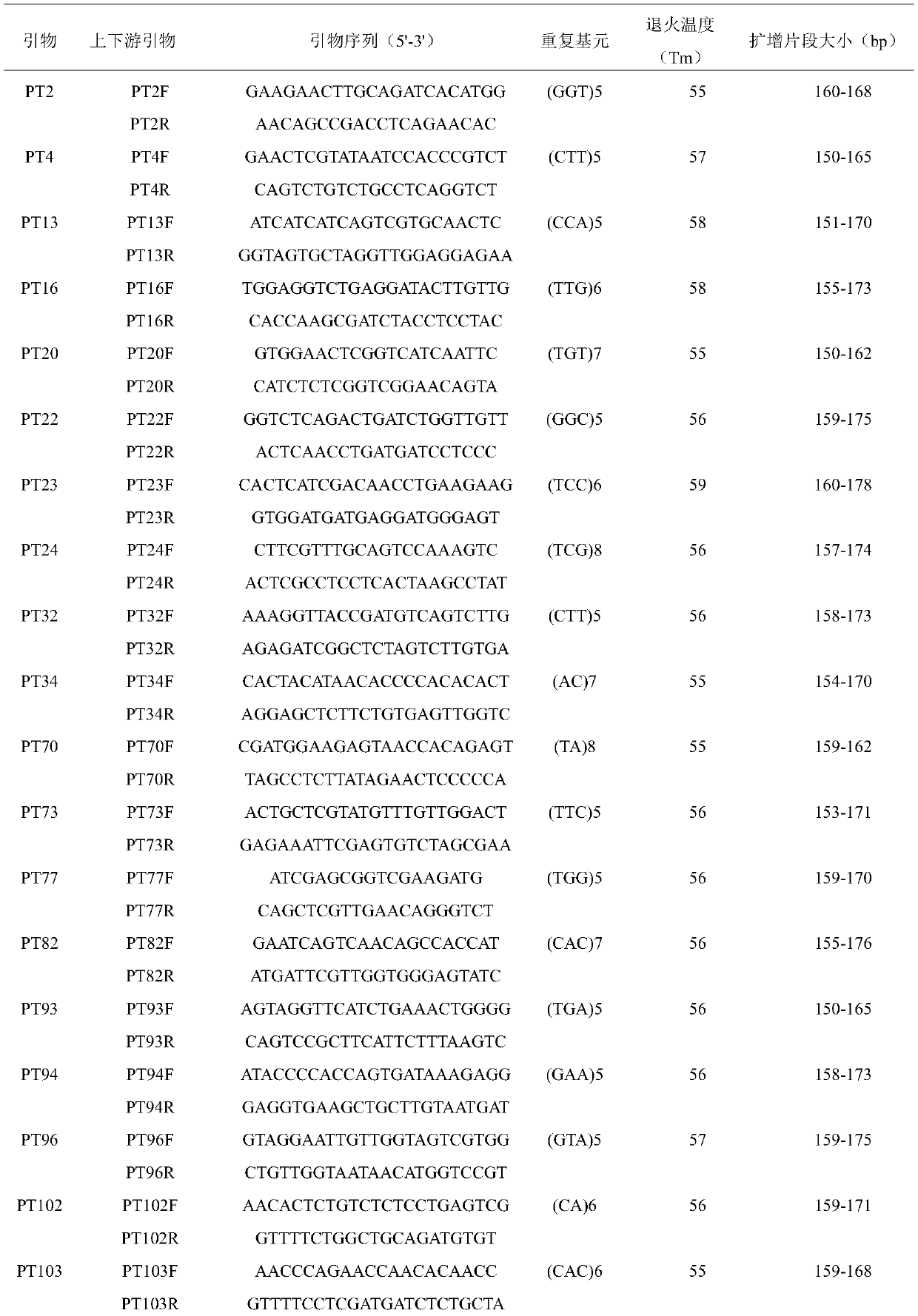
[0043] Through the sequencing and assembly of the Illumina Hiseq2000 platform, 46008 Unigenes were obtained from wheat leaf rust fungus with a N50 of 1064 nt; SSR sites were searched from 46008 Unigenes sequences using MISA software, and a total of 3729 SSR sites were obtained, distributed in 3085. In Unigenes; Primer3 software was used to design primers, 37 pairs of primer sequences were randomly selected and synthesized by Shanghai Shenggong Biological Engineering Co., Ltd. The primer sequence is shown in Table 3.
[0044] Table 3 Sequence information of 37 pairs of primers
[0045]
[0046]
Embodiment 2
[0048] Extraction of Whole Genome DNA of Wheat Leaf Puccinia
[0049] 1) Weigh about 200μg of wheat leaf rust fungus urinifera spores, put them in 2mL centrifuge tubes, put them in a liquid nitrogen kettle, and then fully grind them with a tissue grinder;
[0050] 2) Quickly add 800μL of CTAB solution preheated at 65°C (add 2% mercaptoethanol before use) into the centrifuge tube, put the centrifuge tube in a 65°C water bath for 1h, and mix the sample upside down every 5min during this time;
[0051] 3) After cooling to room temperature, add 800μL of phenol:chloroform:isoamyl alcohol (25:24:1), mix well for 10min, centrifuge at 12000rpm for 15min;
[0052] 4) Aspirate the supernatant, put it into a new centrifuge tube, add 800μL of chloroform: isoamyl alcohol (24:1), mix for 10 minutes, and then centrifuge at 12000rpm for 10 minutes;
[0053] 5) Transfer the supernatant to a new tube, add 1 mL of absolute ethanol pre-cooled at -20°C, mix well, and place it at -20°C for 3 to 4 hours;
[00...
Embodiment 3
[0058] The amplification system is: 25μL total system containing 2μL of template DNA, 1U Taq DNA polymerase, dNTPs 1.0mmol / L, 1.5μL each of the upstream and downstream primers, 1×PCR Buffer, and the rest are supplemented with sterile distilled water to the reaction system to 25.0μL.
[0059] The reaction procedure is: pre-denaturation at 94°C for 5 min, then denaturation at 94°C for 30 seconds, annealing at 55-59°C for 1 min, extension at 72°C for 1 min, 35 cycles, and finally extension at 72°C for 2 min and storage at 10°C.
[0060] The detection method is: take 5μL of the amplified product, add 0.5μL of denatured loading indicator, and separate by electrophoresis in a 10% polyacrylamide gel, the electrophoresis buffer is 0.5×TBE, 300V constant pressure electrophoresis for about 1h, then Silver dyed color.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com