InDel marker based on peanut genome, primer combination and application
A genome and peanut technology, applied in the field of InDel markers based on the peanut genome, can solve the problems of limited number of InDel markers, achieve stable amplification results, protect legitimate rights and interests, and achieve high resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0169] The identification and marker development of embodiment 1 peanut InDel
[0170] 1. Acquisition of peanut genome data
[0171] Download the genome of Tifrunner from the peanut database (https: / / peanutbase.org / ) as a reference genome, and download the sequencing data of Lionhead from the NCBI database (https: / / www.ncbi.nlm.nih.gov / ) ( SRA number: SRR7617991).
[0172] 2. Identification of InDel markers in peanut genome
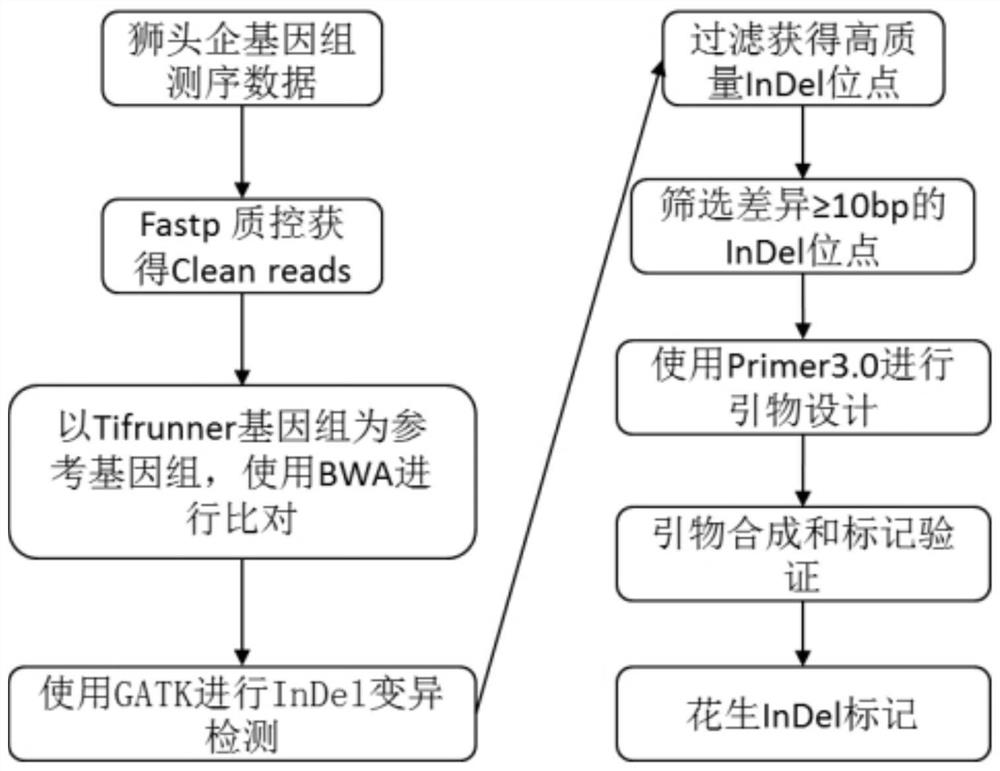
[0173] Such as figure 1 As shown, first use the Fastp software to perform quality control on the obtained lion head enterprise resequencing data to obtain Cleanreads, then use the Tifrunner genome as the reference genome, use the mem program in the BWA software to compare the Cleanreads to the reference genome, and then use the smtools software Perform data format conversion, sort and remove marker PCR repeat sequences, and then use GATK software for variation detection and filtering, and obtain 38,777 InDel sites, and then screen for InDel sites with ...
Embodiment 2
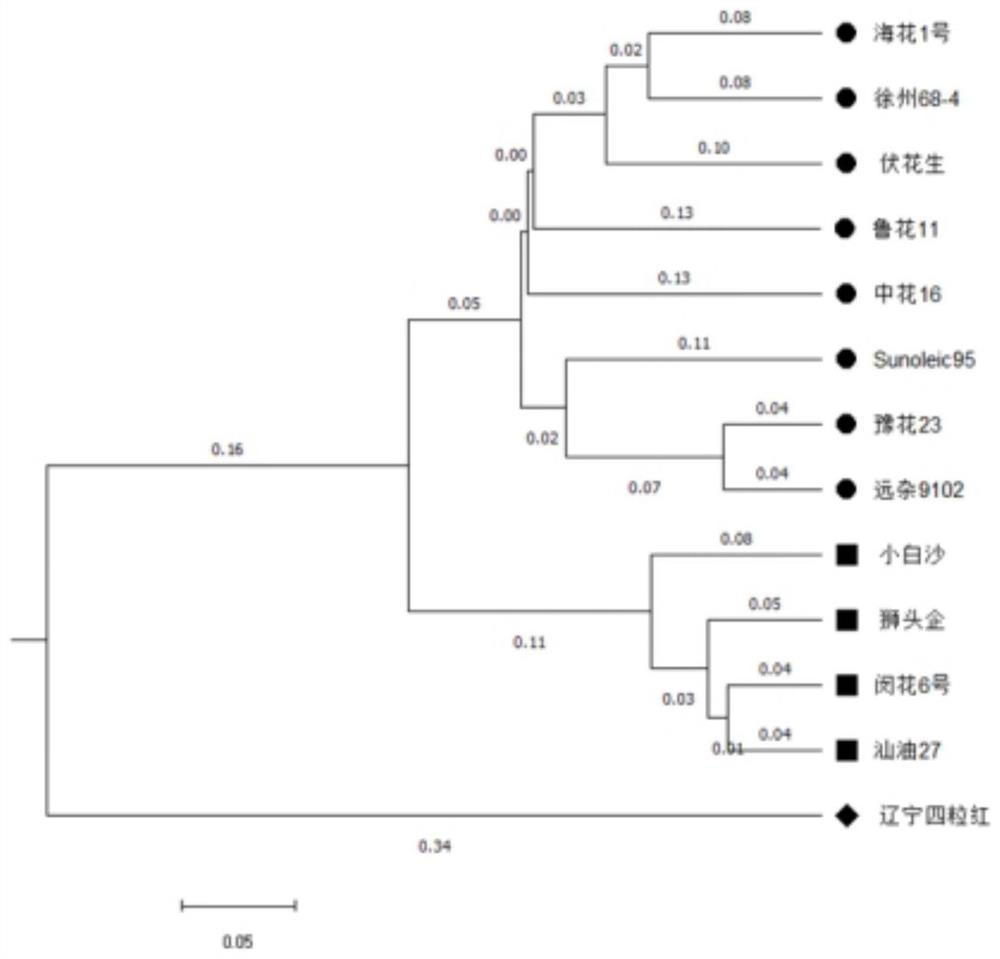
[0184] Example 2 Utilize peanut InDel markers to carry out germplasm analysis
[0185] 1. Polymorphism analysis of InDel markers
[0186] The 13 peanut varieties shown in Table 2 were detected by polypropylene gel electrophoresis using the primers corresponding to the 3615 InDel markers obtained in Example 1.
[0187] (1) Design and synthesize common primers using InDel markers, and then analyze them by polypropylene gel electrophoresis.
[0188] The specific steps are: add 1 / 2 volume of denaturant to the PCR product, mix well, denature at 95°C for 5 minutes, immediately place it on ice to cool, then take 2 μL of the product and use 6% denaturing polyacrylamide gel electrophoresis for detection, nitric acid Silver staining, development, scanning and preservation.
[0189] (2) Organize the experimental data.
[0190] The amplification of peanut InDel markers in 13 peanut varieties was obtained. 100 InDel markers were further identified, and these 100 InDel markers were dist...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com