A kind of SNP marker related to pig backfat thickness and its utilization method
A technology of pigs with thick backfat and breeding pigs, which is applied in the field of pig breeding, can solve the problems of large confidence intervals, etc., and achieve the effects of improving economic benefits, increasing the speed of breeding, and enriching molecular markers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0024] According to a preferred embodiment of the present invention, the SNP marker located at 312, 813, 451 bp of chromosome 1 of the pig reference genome Sscrofa10.2 version sequence has a C / T polymorphism.
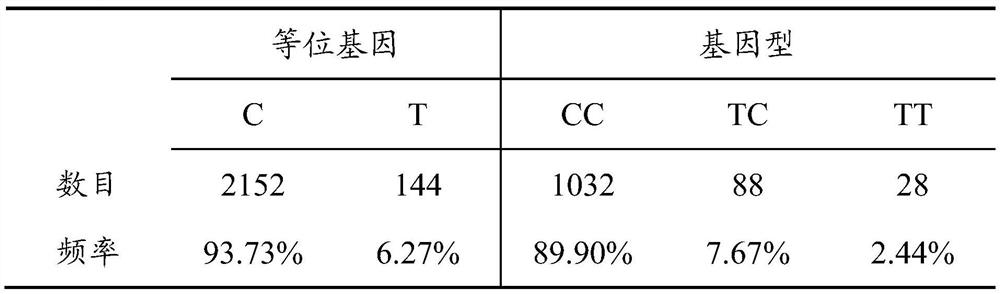
[0025] Among them, the two alleles of the SNP locus are C and T, the gene frequency of C is 0.9773, the gene frequency of T is 0.0627, and C is the dominant allele.
[0026] In a further preferred embodiment, the genotype of the SNP site located at 312, 813, 451 bp of chromosome 1 of the swine reference genome Sscrofa10.2 version sequence is TT genotype pigs have more genotypes than TC and CC pigs Low backfat thickness.
[0027] In the present invention, there are three genotypes corresponding to the SNP site located at the 312, 813, 451 bp of chromosome 1 of the pig reference genome Sscrofa10.2 version sequence, which are TT, TC and CC, respectively, and the TT genotype is that the site is Homozygous for base T, TC genotype is heterozygote for this site, CC genotype i...
Embodiment 1
[0069] Example 1 Acquisition of SNP site
[0070] 1. Experimental group
[0071] The pig population used in this example is 1148 pigs from the basic sow farm of Shenyuan pig in Hebei, including 588 large white pigs, 371 landrace pigs and 189 Duroc pigs.
[0072] 2. Measurement and correction of backfat thickness
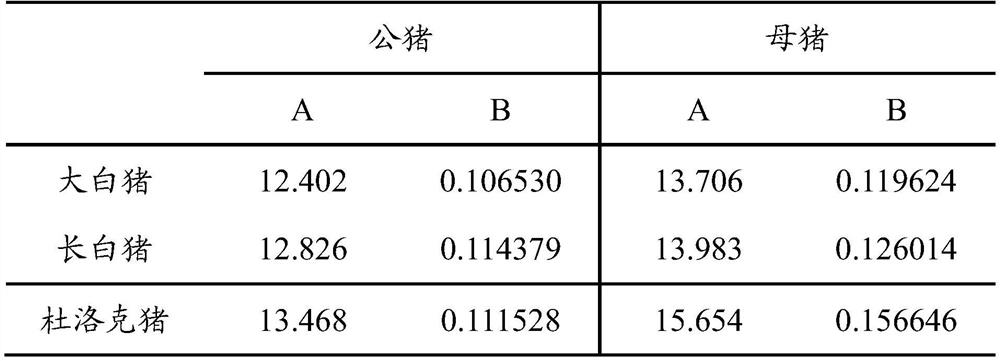
[0073] When the individual body weight of the pig is in the range of 85-105kg, the backfat thickness of the living body is measured, and the backfat thickness at the 3rd to 4th intercostal space from the bottom is measured by B-ultrasound scanning, in millimeters; finally, the weight of pigs up to 100kg is calculated according to the following correction formula The in vivo backfat thickness:
[0074] The relationship between the corrected backfat thickness and the measured backfat thickness is:
[0075] Corrected backfat thickness = measured backfat thickness×CF (wherein: CF=A÷{A+[B×(measured weight-100)]});
[0076] Wherein, sow CF value=(measured body weight...
Embodiment 2
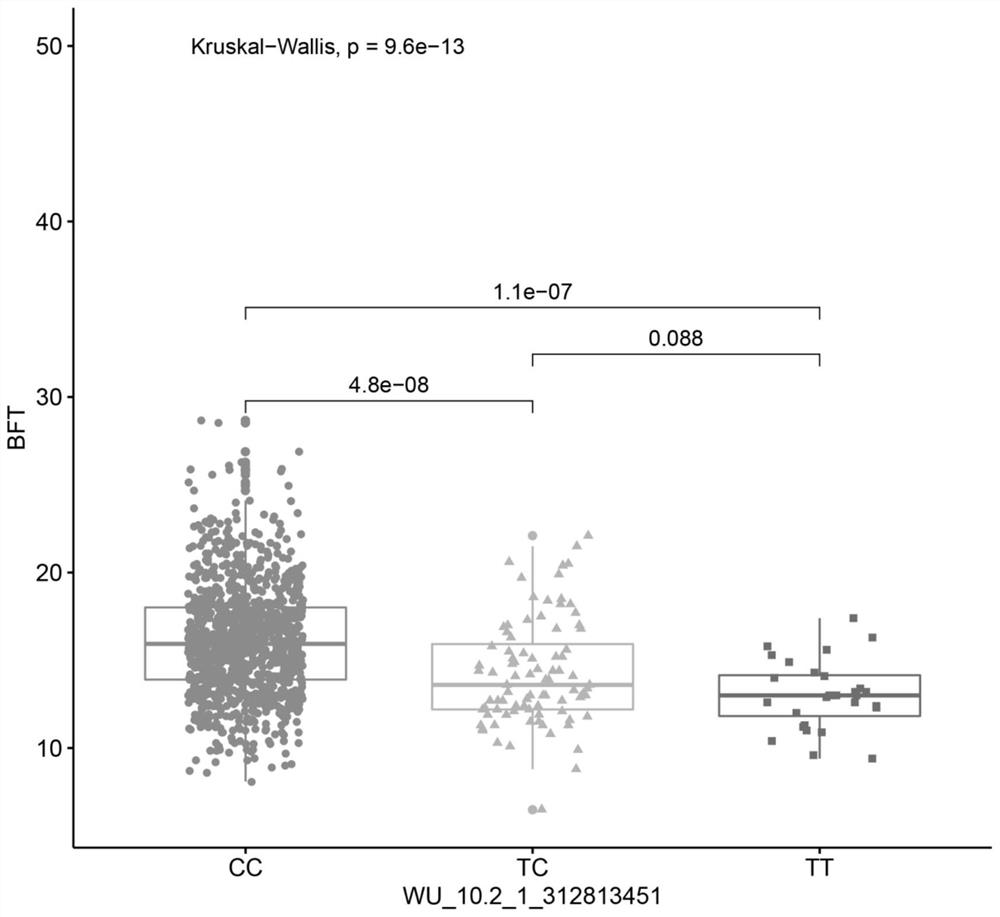
[0100] Example 2 Correlation results between different genotypes of SNP loci and backfat thickness
[0101] Take the genomic DNA of the pig population in Example 1, use Neogen_POR80K of Neogen Corporation to detect the genotype of each individual, extract the sequence of the polymorphic site by R language, and count the genotype frequency and gene frequency distribution of the SNP site, The results are shown in Table 2:
[0102] Table 2
[0103]
[0104] It can be seen from Table 2 that C is the dominant allele, and the CC genotype is the dominant genotype of the experimental population.
[0105] The Kruskal-Wallis method was used to test the significance of differences between genotype data and phenotype data using Rstudio software, where P-value<0.05 indicated a significant difference. The correlation analysis results are shown in Table 3:
[0106] table 3
[0107]
[0108] It can be seen from Table 3 that the corrected backfat thickness of the three genotypes of t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com