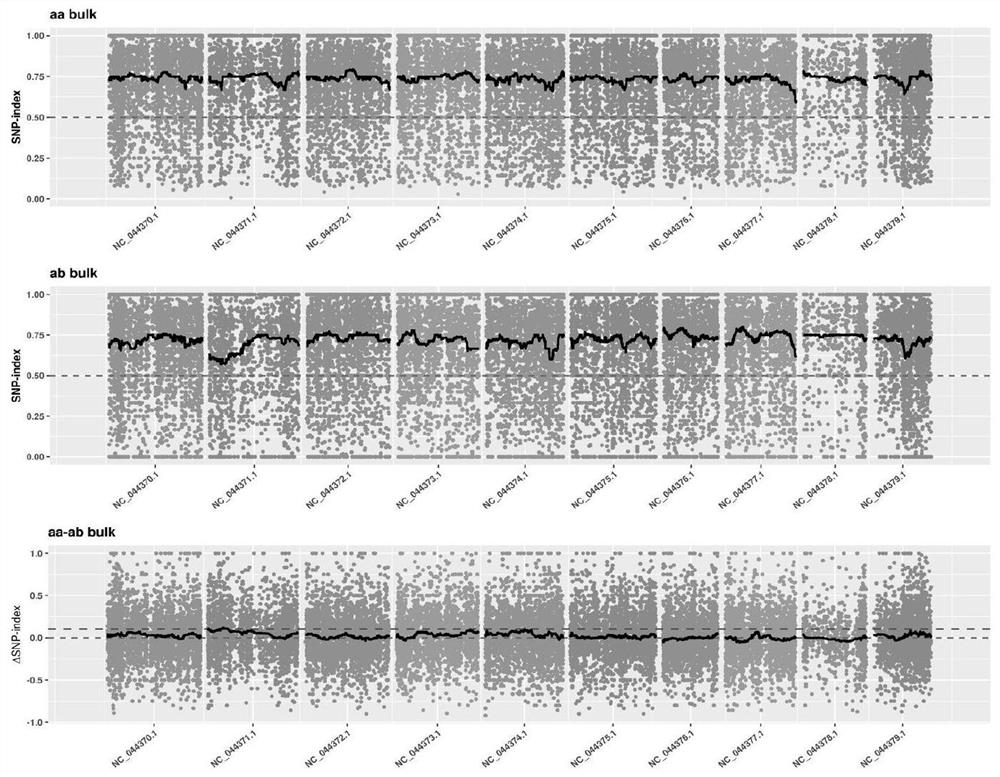
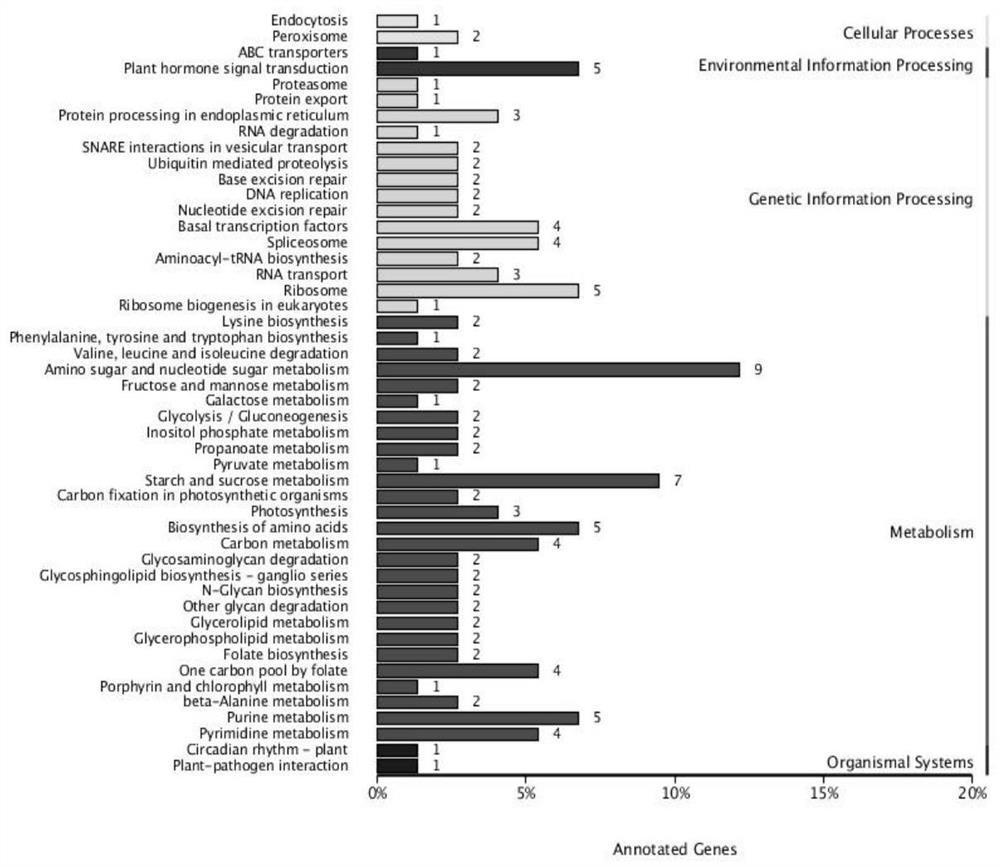
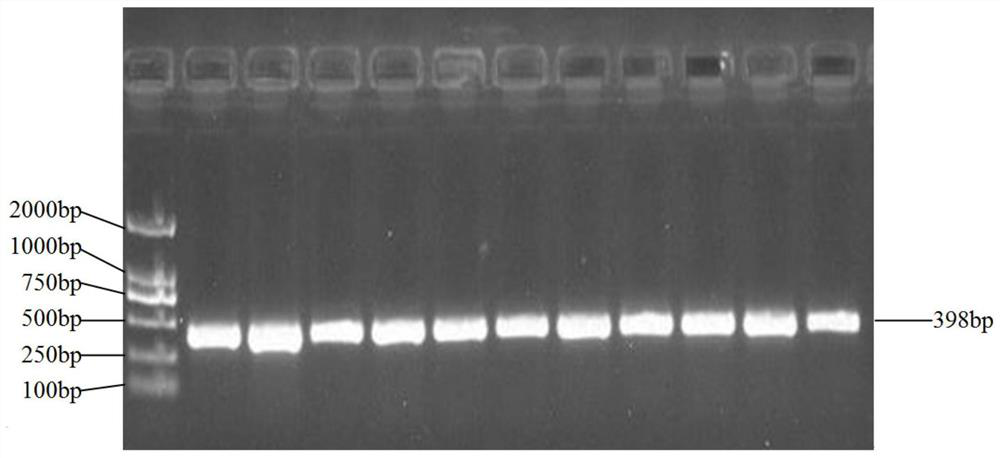
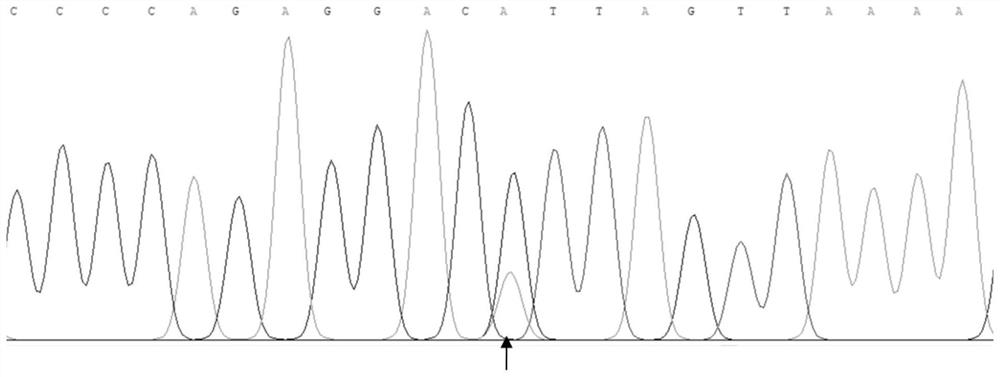
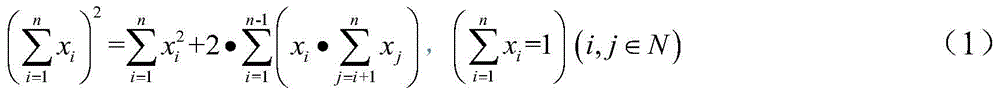Patents
Literature
32 results about "Genotype frequency" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Genetic variation in populations can be analyzed and quantified by the frequency of alleles. Two fundamental calculations are central to population genetics: allele frequencies and genotype frequencies. Genotype frequency in a population is the number of individuals with a given genotype divided by the total number of individuals in the population. In population genetics, the genotype frequency is the frequency or proportion (i.e., 0 < f < 1) of genotypes in a population.
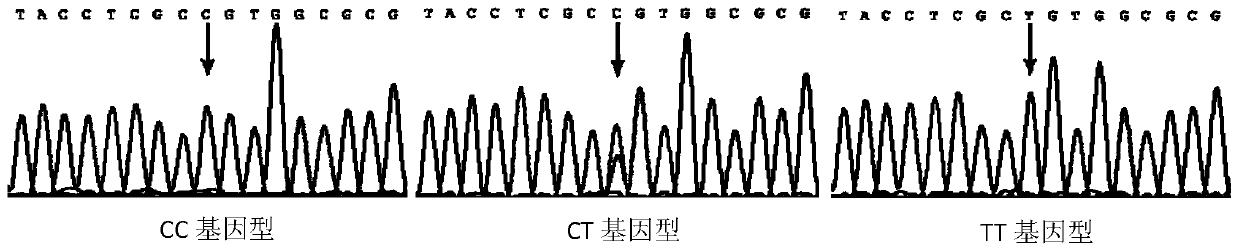
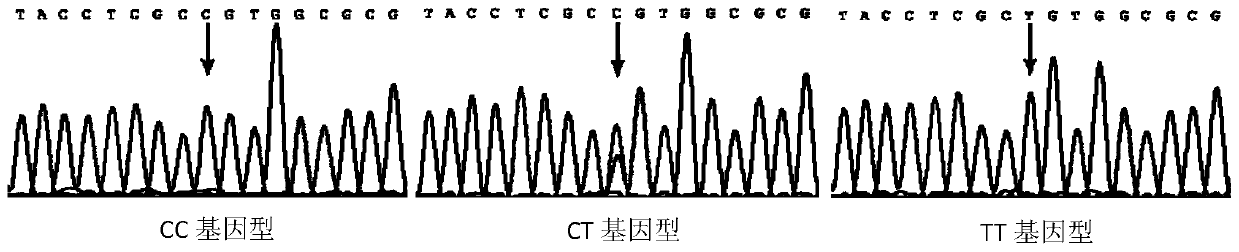
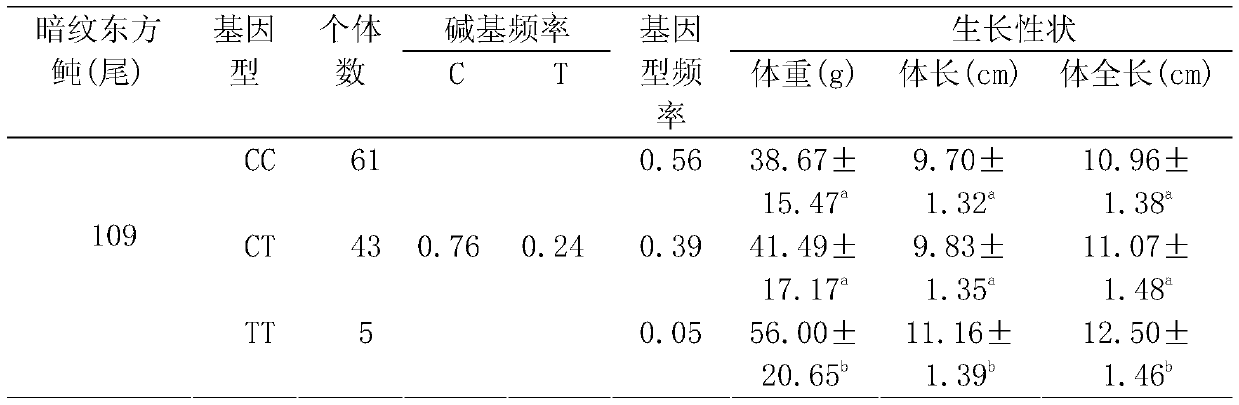
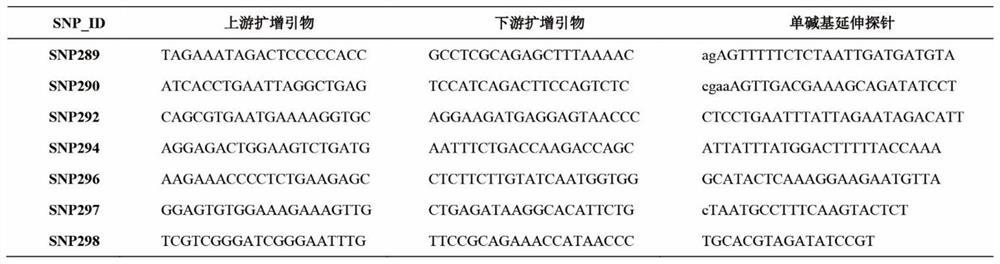
SNP locus related to rapid growth of takifugu obscurus and application thereof
ActiveCN110004235AMicrobiological testing/measurementClimate change adaptationNucleotideTakifugu obscurus
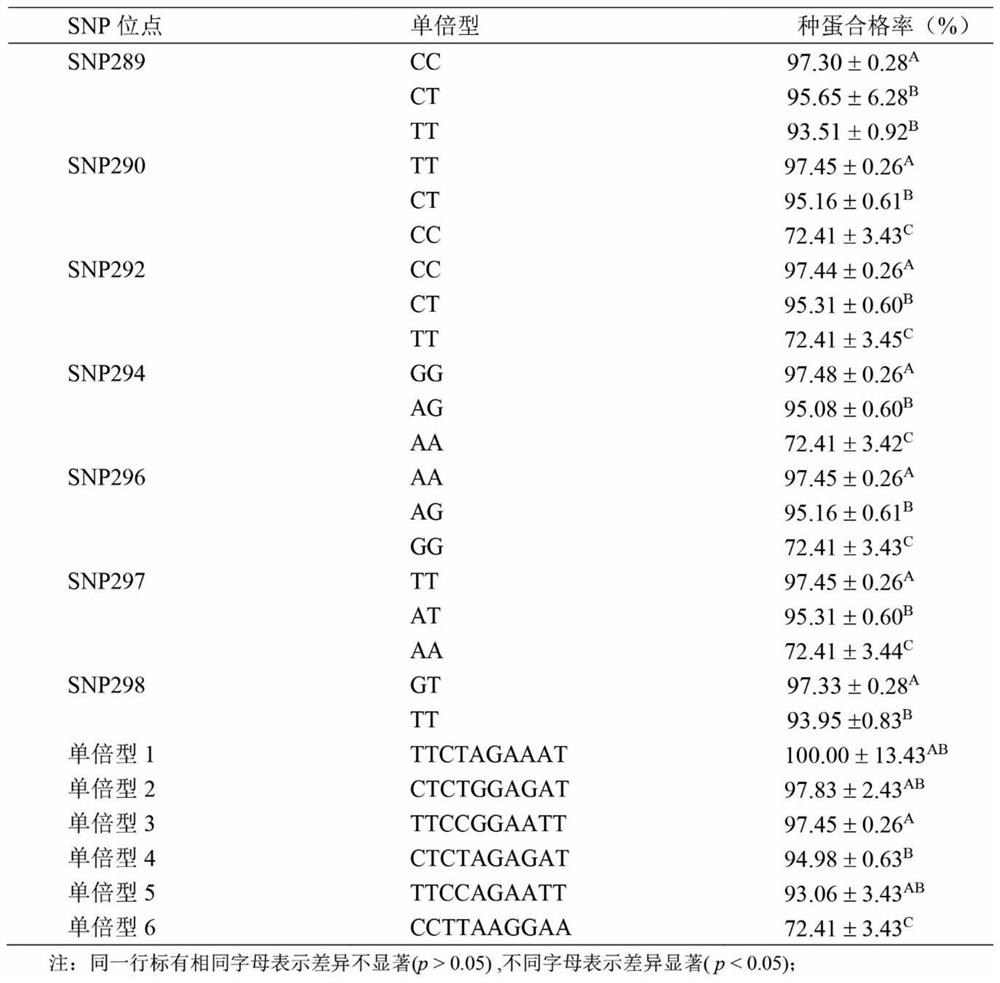
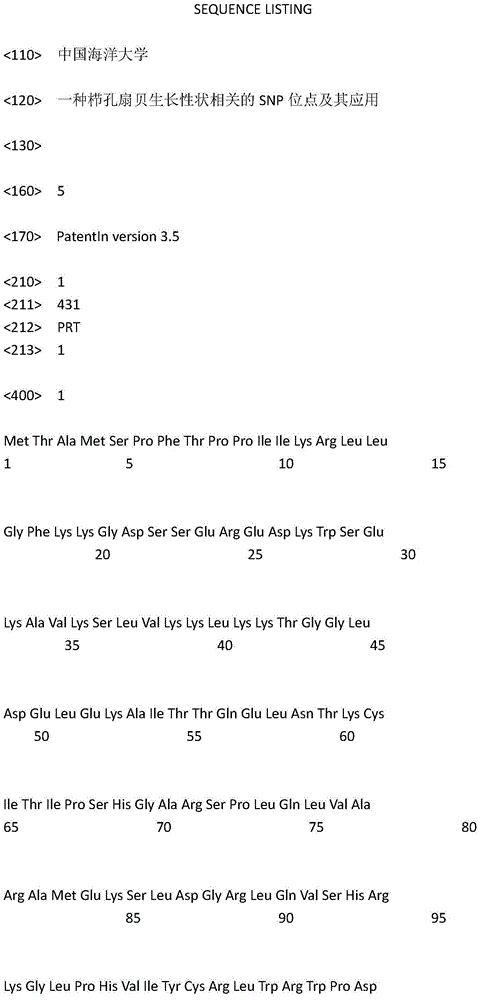
The invention provides an SNP locus related to rapid growth of takifugu obscurus and application. The SNP locus is located at position 724 of an MSTN gene with a nucleotide sequence shown as SEQ ID NO:1, and the base of the SNP locus is C or T. The SNP locus is used for breeding takifugu obscurus individuals with rapid growth potential. According to the method, by analyzing the correlation betweenthe locus genotype frequency and the growth traits of the takifugu obscurus, the SNP locus related to the growth traits is found at 724 base position of the MSTN gene of the takifugu obscurus, and the body weight, the body length and the body full length of individuals with TT homozygotic genotypes are obviously greater than phenotypic values of the growth traits of individuals with CC and CT genotypes (p smaller than 0.05). Thus, individuals with TT genotype at the locus can be preferentially selected as parents or for large-scale breeding in production.
Owner:DALIAN OCEAN UNIV
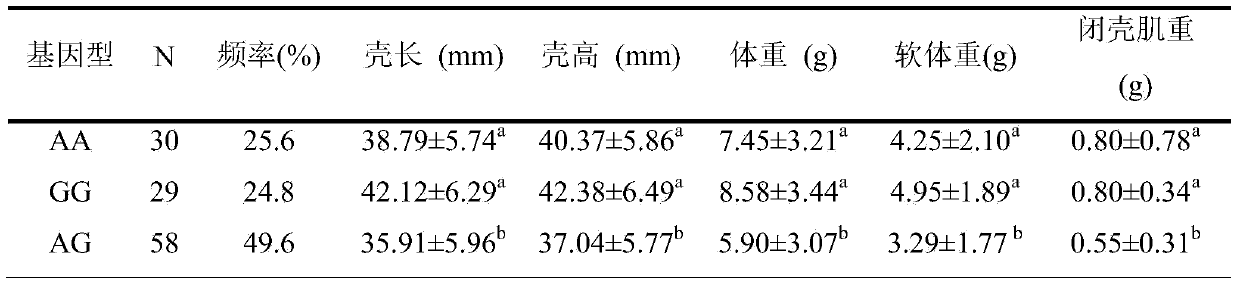
SNP locus related to growth characteristics of patinopecten yessoensis and detection and application thereof
InactiveCN103740729AHigh trait valueAvoid crossbreedingMicrobiological testing/measurementClimate change adaptationInsulin-like growth factor bindingNucleotide sequencing
The invention provides an SNP locus related to growth characteristics of patinopecten yessoensis. The locus is the 1054 site of IGFBP (Insulin Like Growth Factor Binding Proteins) genes of which the nucleotide sequence is shown as SEQ ID NO:2 and has the base of A or G. The invention also provides a probe for detecting the SNP locus and a parting primer. The transcription sequence of the IGFBP genes in the patinopecten yessoensis is subjected to sequencing and Clustal comparison, and three SNP loci are screened. The site polymorphism is detected in the patinopecten yessoensis group by using a high resolution melting curve technology, and correlation between the site genotype frequency and the growth characteristics of patinopecten yessoensis is analyzed. The locus C.1054A>G is obviously related to important growth characteristics such as the height, shell length, weight, soft weight and adductor muscle weight of the patinopecten yessoensis, the growth characteristics of the AG type individuals are obviously lower than those of AA and GG type individuals (P is less than 0.05), and the characteristic value of the GG type is the highest. Therefore, individuals of which the genotype is GG on the locus can be preferentially selected during production and are used as parents for performing high-yield patinopecten yessoensis breeding or performing large-scale breeding.
Owner:OCEAN UNIV OF CHINA
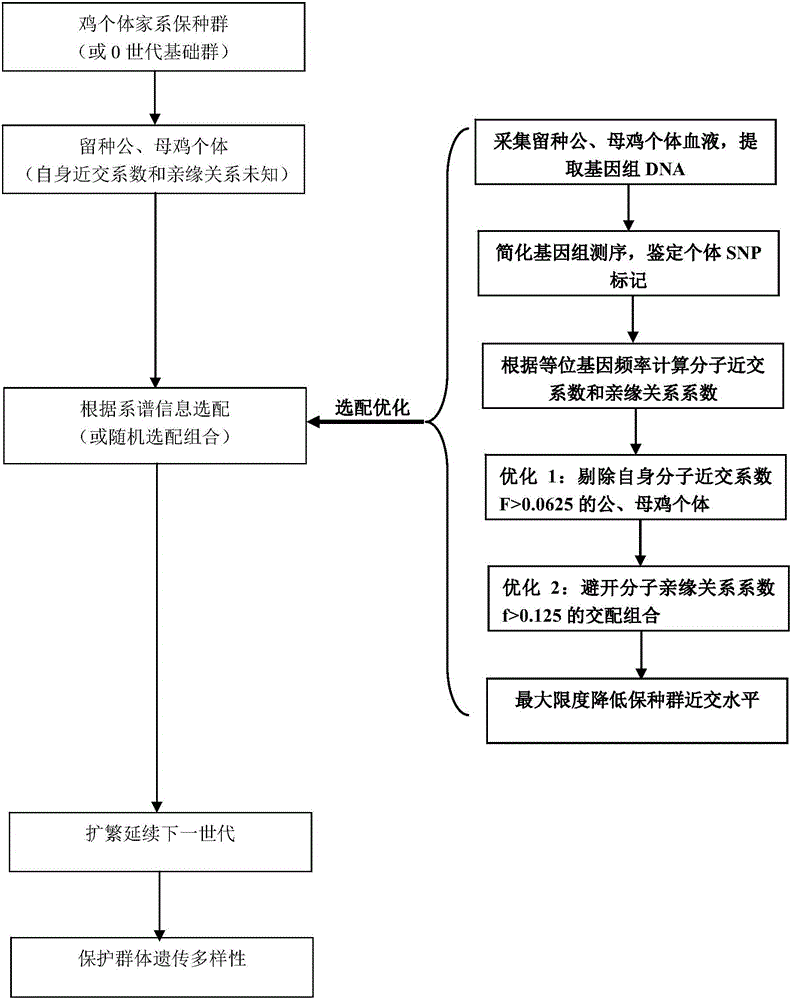
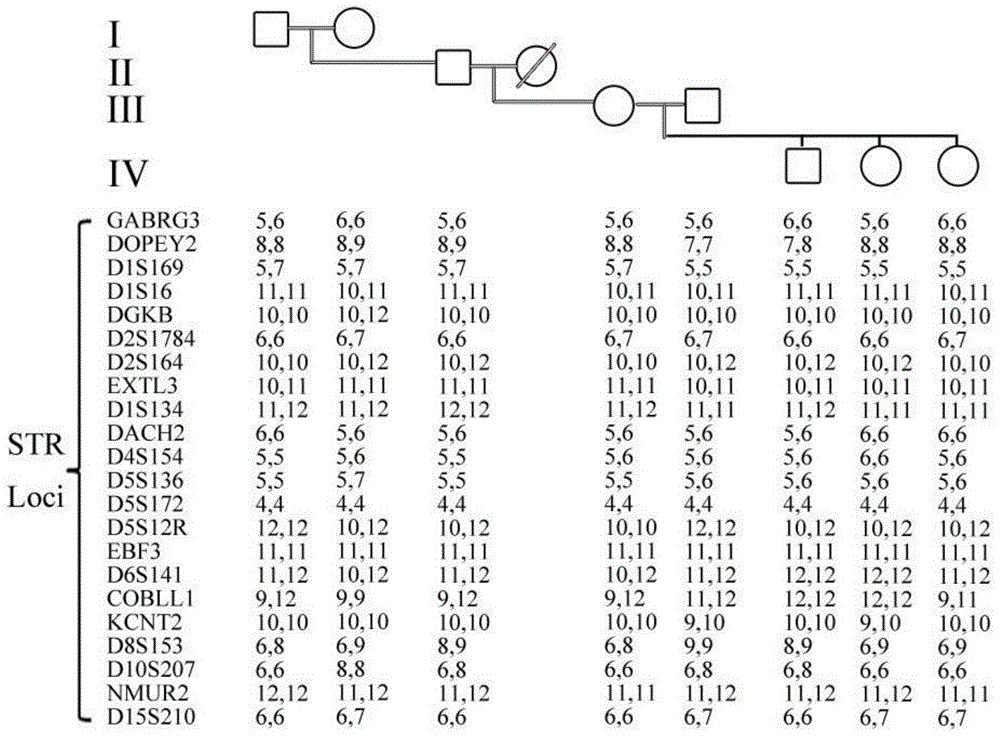
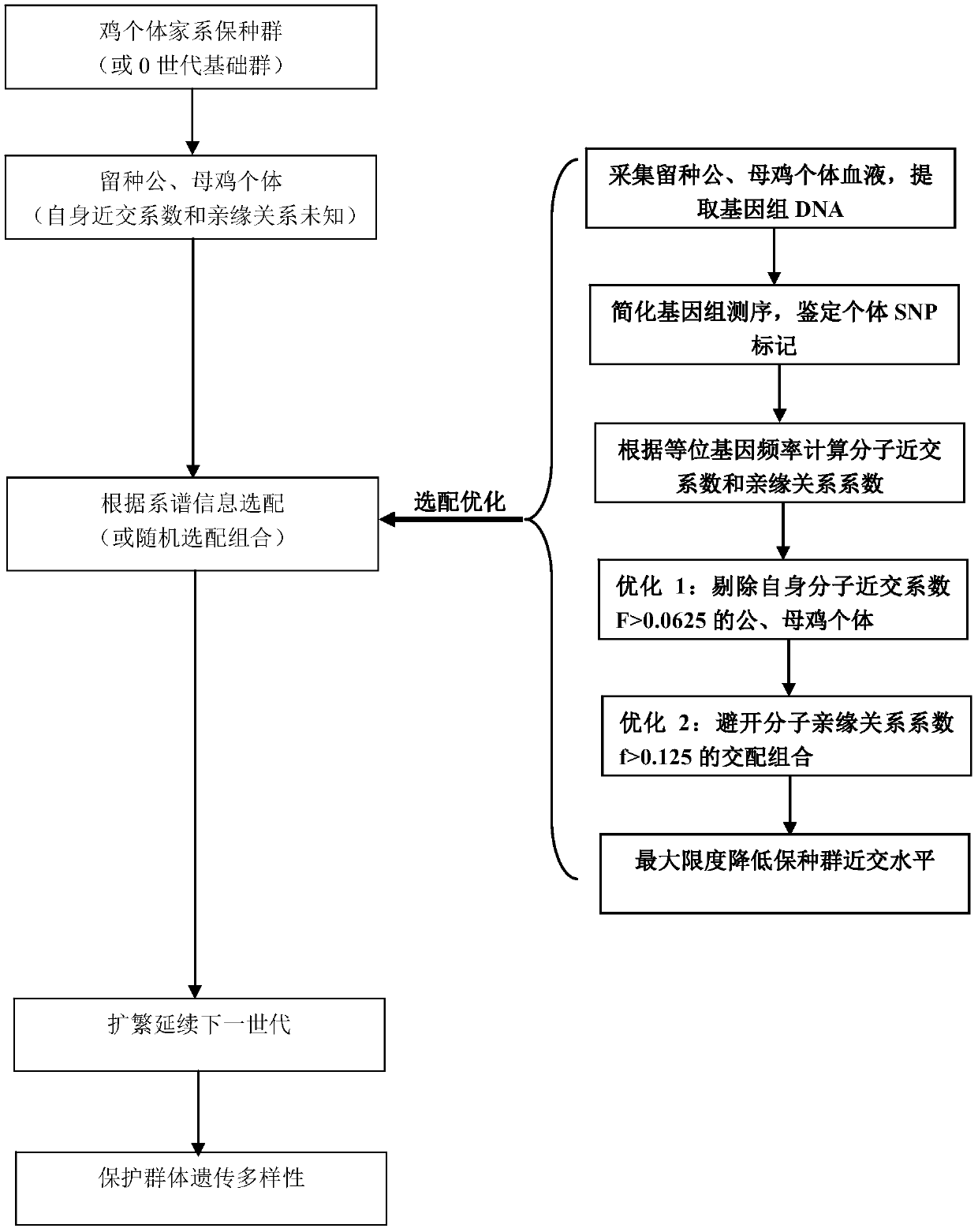
Chicken preserved population individual apolegamy optimizing method
ActiveCN106086172AReduce the level of inbreedingSolve the problem that individual fine matching cannot be carried outMicrobiological testing/measurementGenomic sequencingGenetic diversity
The invention discloses a chicken preserved population individual apolegamy optimizing method. Blood DNA of chicken preserved population breed reserving cock and hen individuals is extracted, sequencing is carried out through a reduced genomic sequencing technology, and the sequence is compared with a sequence of the GenBank database reference chicken breed to find SNP loci existing in different individuals. The individual own inbreeding coefficient and the interindividual phylogenetic relationship coefficient are calculated through gene frequency and genotype frequency of the SNP loci, individual apolegamy is optimized according to the statistics value, mating combination of individuals with an overlarge inbreeding coefficient and individuals with an overlarge within-family phylogenetic relationship coefficient is avoided, and the purposes of reducing the preserved population inbreeding level and maintaining the genetic diversity total quantity steady state are achieved. By means of the method, the problem that fine individual apolegamy cannot be carried out depending on genealogy information in the chicken breed resource protection process can be effectively solved, the preserved population inbreeding level can be effectively reduced, the genetic diversity steady state is maintained, and the breed preservation effect and efficiency are improved.
Owner:JIANGSU INST OF POULTRY SCI
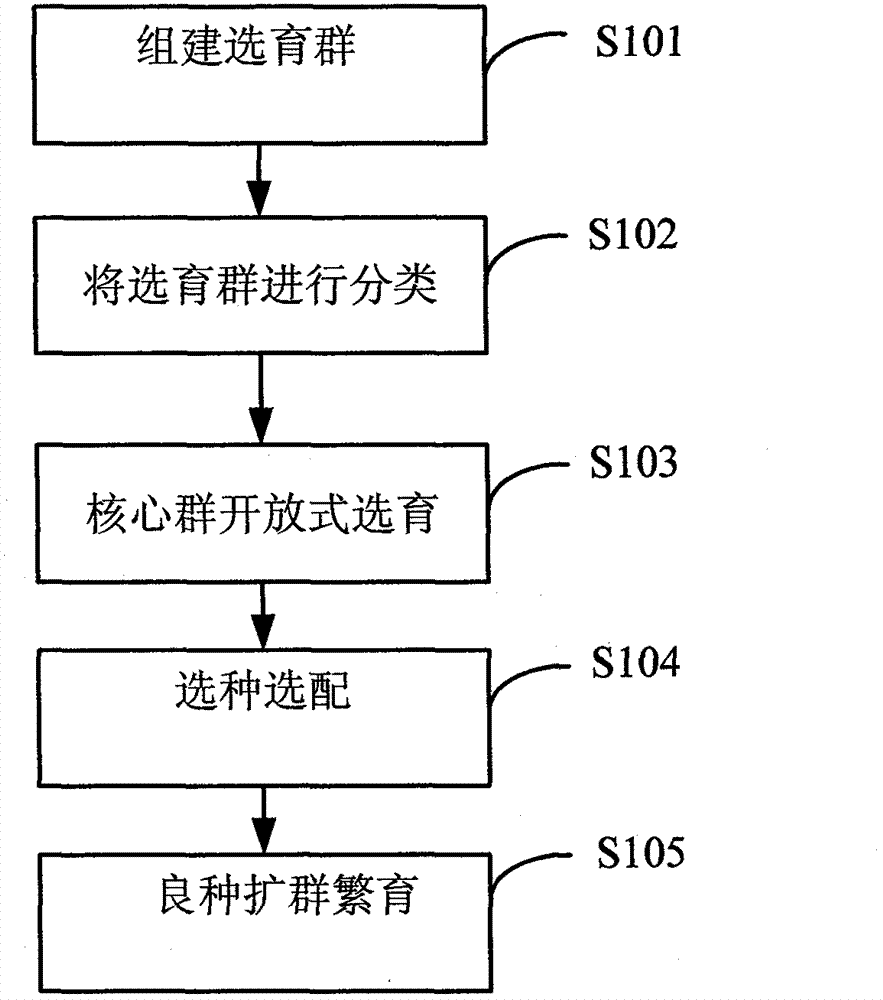
Purification and rejuvenation method of sheep and goat breeds
The invention discloses a purification and rejuvenation method of sheep and goat breeds. The purification and rejuvenation method adopts a core group open type breeding method for establishing a core group and an underlying group among peasant households and realizes the transform from a small scale of the core group of conventional field construction, breed protection and breeding to a large scale of the core group of open type breeding established among the peasant households, fully reflects the breeding advantages of large core group, high frequency of elite genotypes, wide influence surface and wide radiation scope, and improves the production level of the breed protection and breeding group faster; the purification and rejuvenation method realizes the transform from the conventional field construction, breed protection and breeding with high maintenance cost and difficulty in long-term preservation of the core group to the dispersed establishment of the core group among the peasant households, with easiness in maintenance, long-term preservation and consolidation, fully reflects the advantages of large group and small breeding risk, and enables the development of the breed protection and breeding group to have a solid foundation; and the inbreeding coefficient of the open type breeding is slow in increase and difficult to cause the inbreeding depression phenomenon.
Owner:BIJIE INST OF ANIMAL HUSBANDRY & VETERINARY SCI
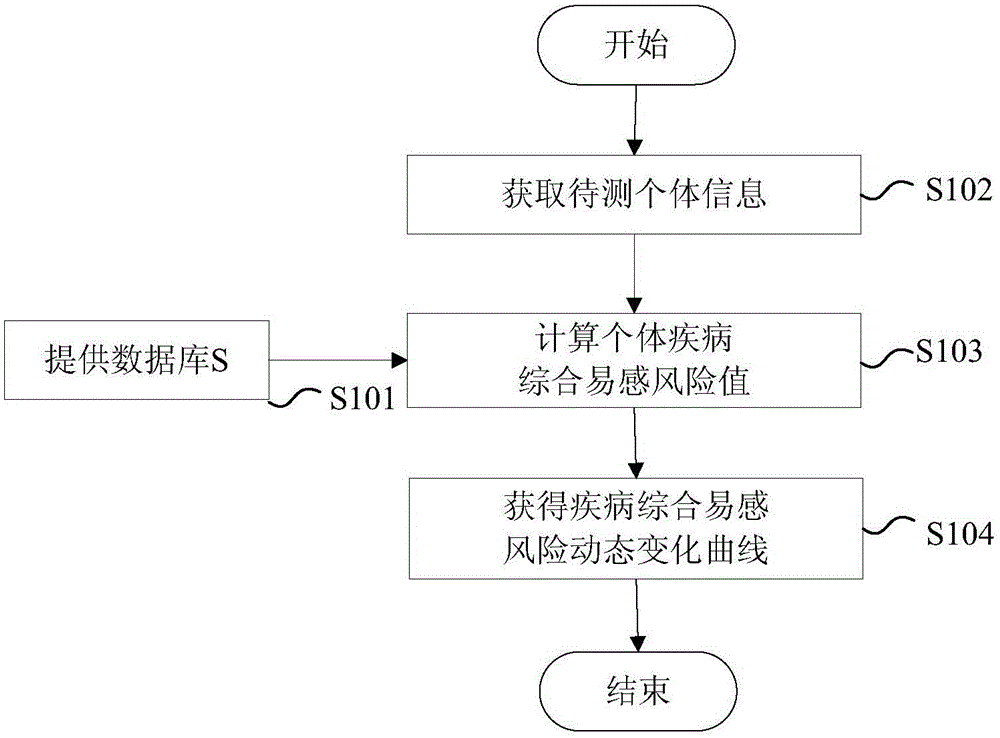
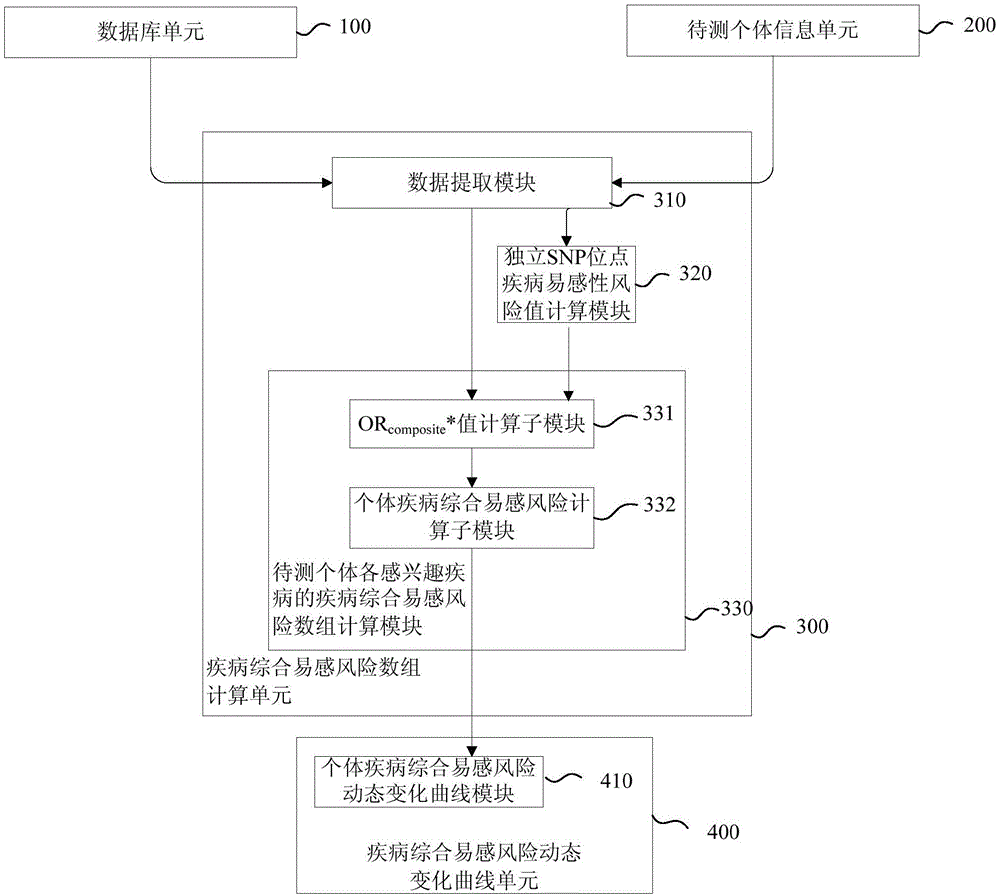
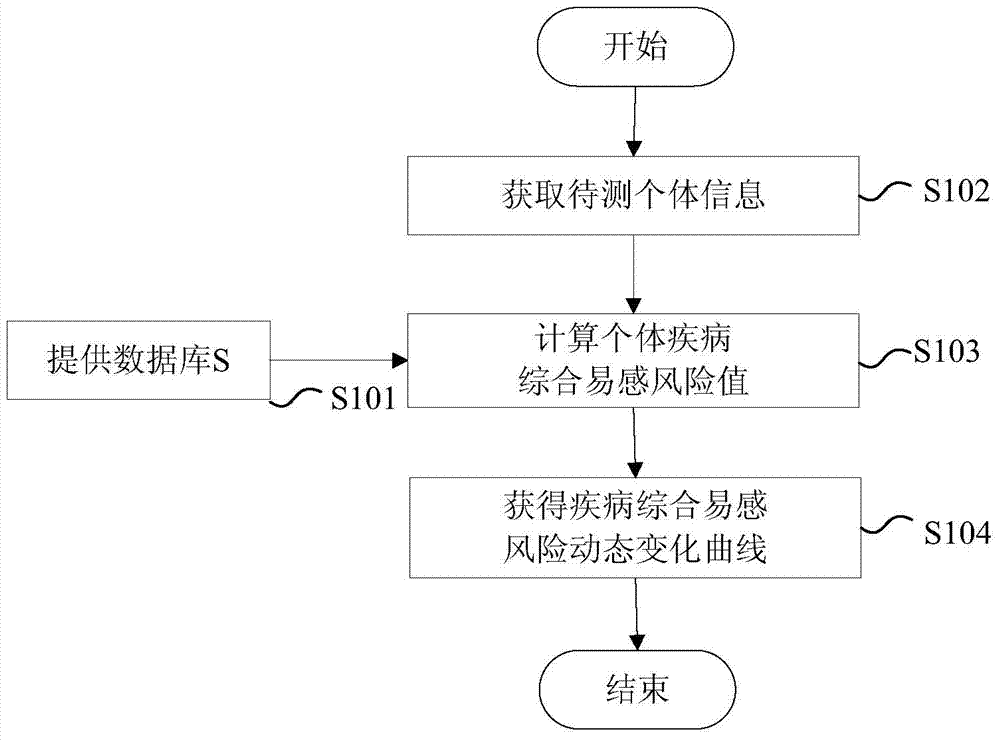
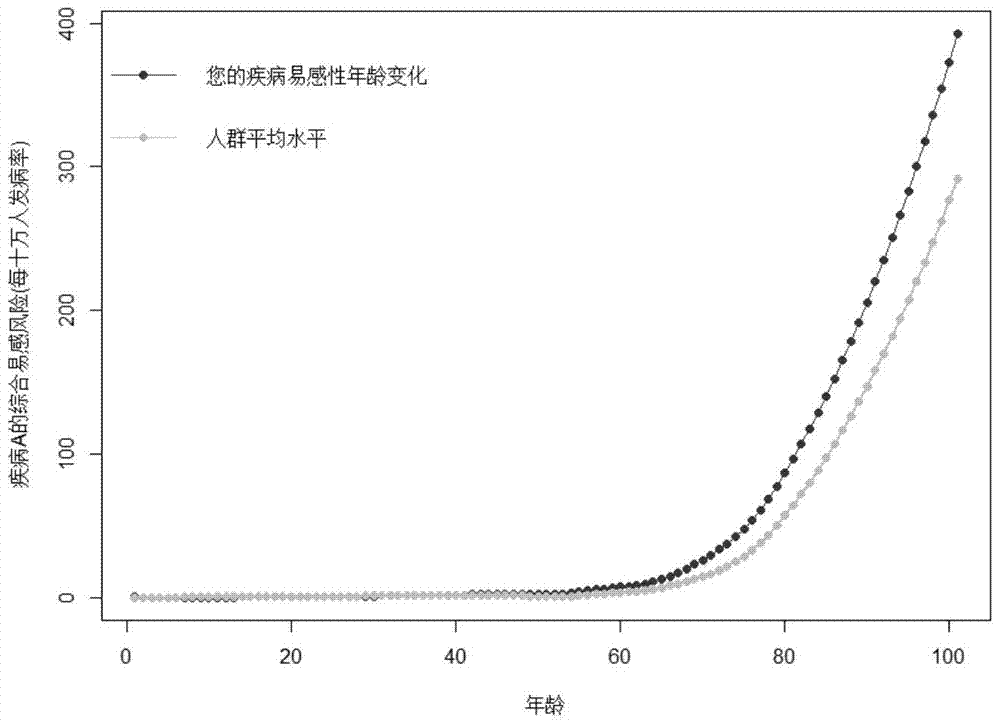
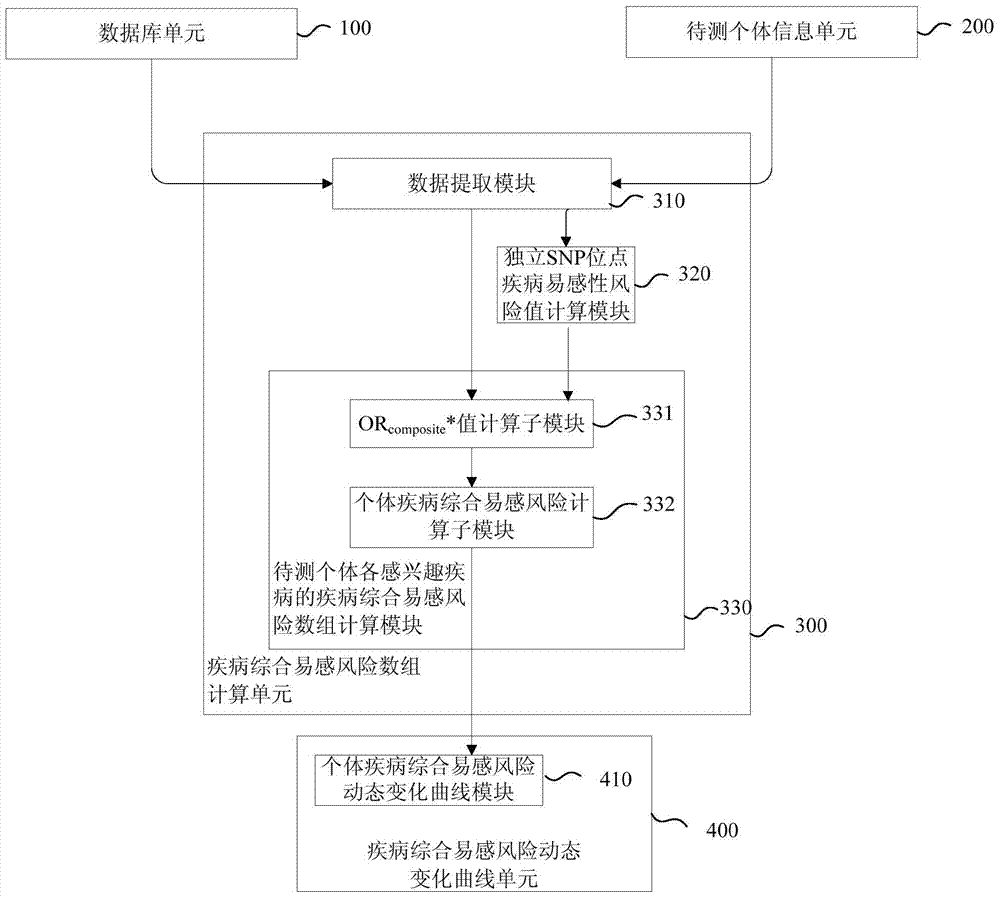
Disease susceptibility risk prediction method and device
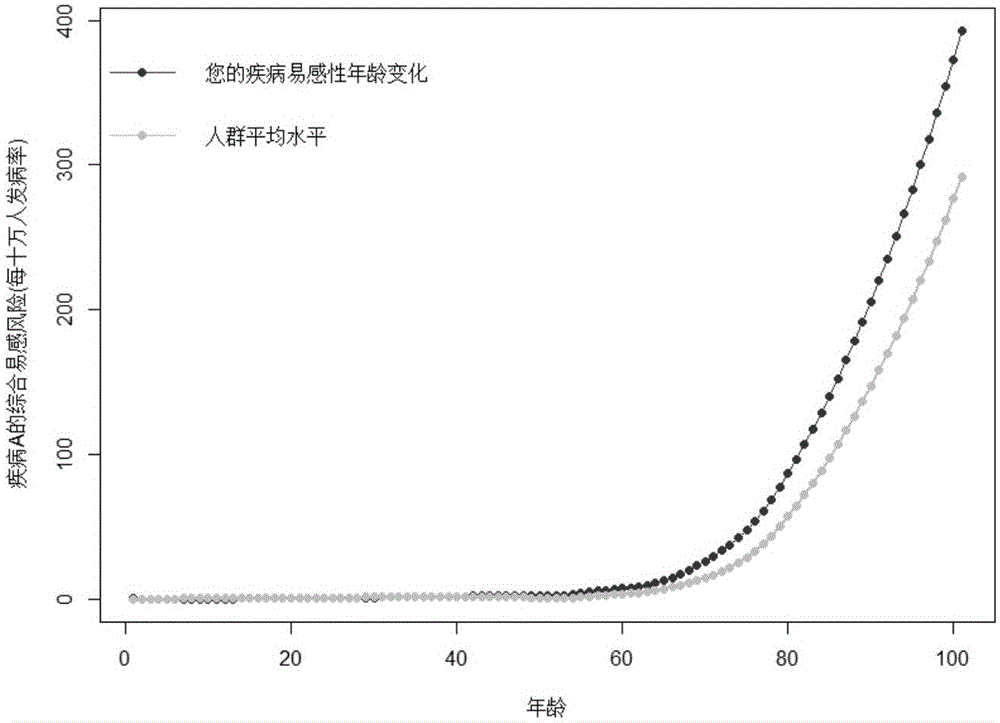
ActiveCN105069322AHelp with health managementTo achieve the effect of "preventive cure"Special data processing applicationsHomozygous genotypeGenotype frequency
The invention relates to bioinformatics and provides a disease susceptibility risk prediction method and device. The disease susceptibility risk prediction method comprises the steps: providing a database including disease morbidity data, SNP locus genotype frequency data, and risk allele homozygous and heterozygous genotype OR value data aiming at each disease-associated SNP locus; receiving information of a to-be-predicted individual; calculating to obtain a comprehensive disease susceptibility risk array of diseases interested by the to-be-predicted individual; and generating a comprehensive disease susceptibility risk dynamic change curve of the individual in an appointed age range. According to the disease susceptibility risk prediction method and device, two factors , including individual inheritance and environment factors, are considered at the same time when the disease susceptibility risk of an individual is calculated, so that calculating results greatly conform to objective reality; according to the obtained disease susceptibility risk age change curve of the individual, the individual not only can learn more precise immediate disease susceptibility risk, but also can continuously understand the susceptibility change tendency of various diseases along with the growth of age.
Owner:上海尔云信息科技有限公司
Method and system used for deducing Han, Tibetan or Wei population source of individual with unknown source
ActiveCN103146820AEasy to classifyImprove accuracyMicrobiological testing/measurementUnknown SourceGenotype
The invention provides a method and a system used for deducing the Han, Tibetan or Wei population source of an individual with an unknown source. The method comprises the following steps: detecting DNA of the individual with an unknown source to obtain genotypes of 94 specific sites of Han, Tibetan and Wei populations; acquiring genotype frequencies of each specific site of the individual with an unknown source in Han, Tibetan and Wei populations respectively; multiplying the genotype frequencies of each specific site to obtain coupling probability values of the 94 specific sites in Han, Tibetan and Wei populations respectively; and determining a population with a highest coupling probability value as a source population of the individual with an unknown source. With the method and the system in the invention, high accuracy deduction of a Han, Tibetan or Wei population source can be realized.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
Paralichthys olivaceus fertility related SNP molecular marker and screening method and application thereof
ActiveCN109385482AEasy to prepareVariety of mutationsMicrobiological testing/measurementRe sequencingScreening method
The invention discloses a paralichthys olivaceus fertility related SNP molecular marker and a screening method and application thereof. The screening method comprises the following steps: separately carrying out cyp21a gene re-sequencing on fertile paralichthys olivaceus and infertile DH paralichthys olivaceus at first to preliminarily screen difference SNP sites; verifying the SNP sites on a large scale by using a fertile population capable of spawning normally in a breeding season and DH infertile population incapable of spawning; carrying out parting on the SNP sites, which are obtained instep (1), of the paralichthys olivaceus by using a multiple SNaPshot SNP parting method; and carrying out statistics on genotype frequency of the SNP sites which are successful in parting by using SPSS software, and carrying out difference significance analysis by using chi-square test to screen out SNP sites related to paralichthys olivaceus fertility. In the invention, a paralichthys olivaceus fertility research model is established, a method for rapidly establishing a large number of 21OHD animal models is provided, and SNP sites of fertility related cyp21a genes are obtained by screening.
Owner:中国水产科学研究院北戴河中心实验站
Application of anti-WSSV candidate molecular marker of nLvALF1 gene of Litopenaeus vannamei to genetic breeding of Litopenaeus vannamei
InactiveCN106011252AEasy to operateShort cycleMicrobiological testing/measurementDNA/RNA fragmentationDiseaseCorrelation analysis
The invention relates to application of an anti-WSSV candidate molecular marker of the nLvALF1 gene of Litopenaeus vannamei to genetic breeding of Litopenaeus vannamei, belonging to the field of genetic breeding of aquatic livestock. Through preliminary study, three SNP markers (P<0.05) significantly correlated with anti-WSSV are screened out from the nLvALF1 gene by using a correlation analysis method. In the invention, resistance lines and non-resistance lines of the three SNP markers in other genetic breeding materials are subjected to SNP genotyping so as to eventually screen out one SNP marker, located at a site nLvALF1g-114-T>C, significantly correlated with anti-WSSV; the genotype frequency of the CC type of the site in resistance lines is substantially higher than the genotype frequency of the CC type of the site in non-resistance lines; and a method for molecular marker-aided genetic breeding of anti-WSSV is established by using the marker. The invention provides a candidate technical means for molecular marker-aided breeding for disease resistance and lays foundation for research on breeding of an anti-WSSV variety of Litopenaeus vannamei.
Owner:INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Method for detecting single nucleotide polymorphism of SPATA6 gene of sheep and application
ActiveCN110241227APromote amplificationEasy to unlinkMicrobiological testing/measurementProteomicsAnalysis dataNucleotide
The invention discloses a method for detecting single nucleotide polymorphism of a SPATA6 gene of sheep. The method comprises the following steps of S1, selecting samples; S2, constructing a cDNA pond; S3, performing PCR amplification; S4, performing product sequencing and single nucleotide polymorphism subtyping; and S5, performing frequency statistics of single nucleotide polymorphism sites of the SPATA6 gene of the sheep and analyzing the correlation between the frequency statistics and testis phenotypes. The invention further provides an application of the detected nucleotide polymorphism of the SPATA6 gene of the sheep. Through genotypic frequency calculation, the subtyping result of a fluorescence multiple enzyme connection reaction technique, testis phenotype data and SAS(9.2) software analysis data, the relationship between the testis phenotype and the single nucleotide polymorphism mutated by the 72272nd T>C of the spermatogenesis related gene 6 is sufficiently constructed, and high-fecundity sheep are bred for screening and labeling.
Owner:甘肃润牧生物工程有限责任公司
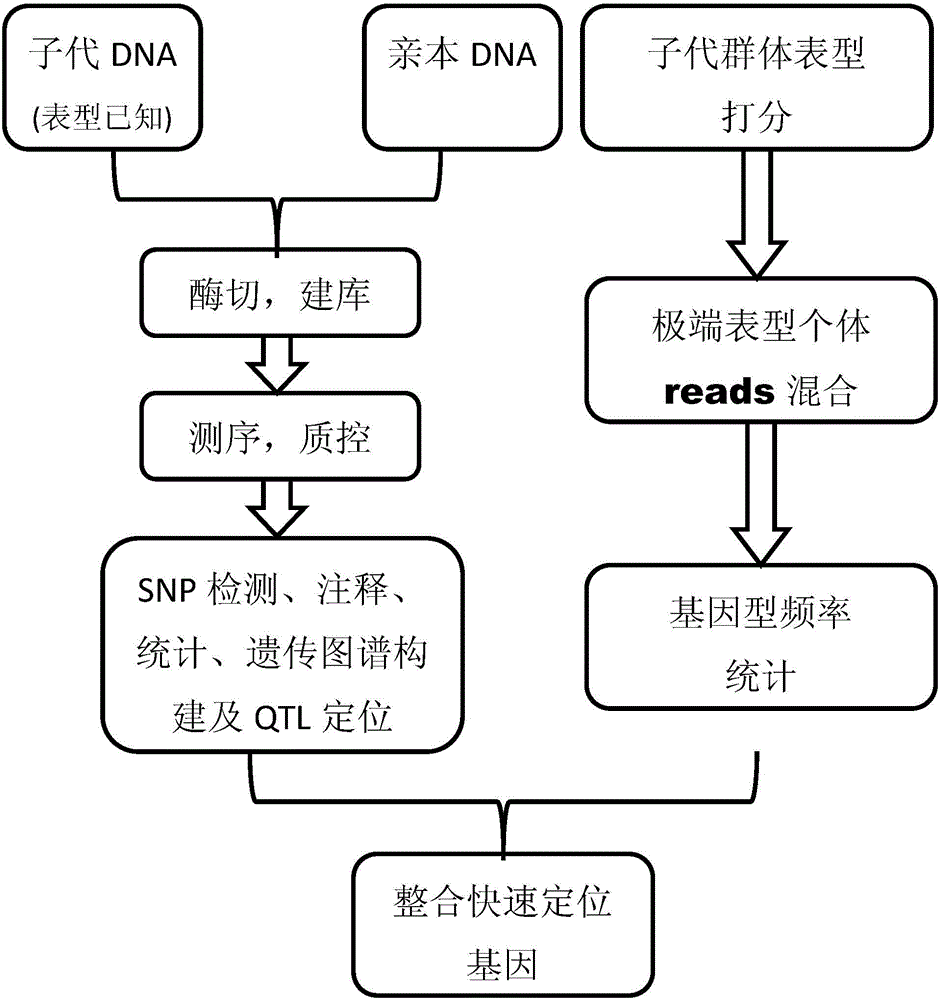
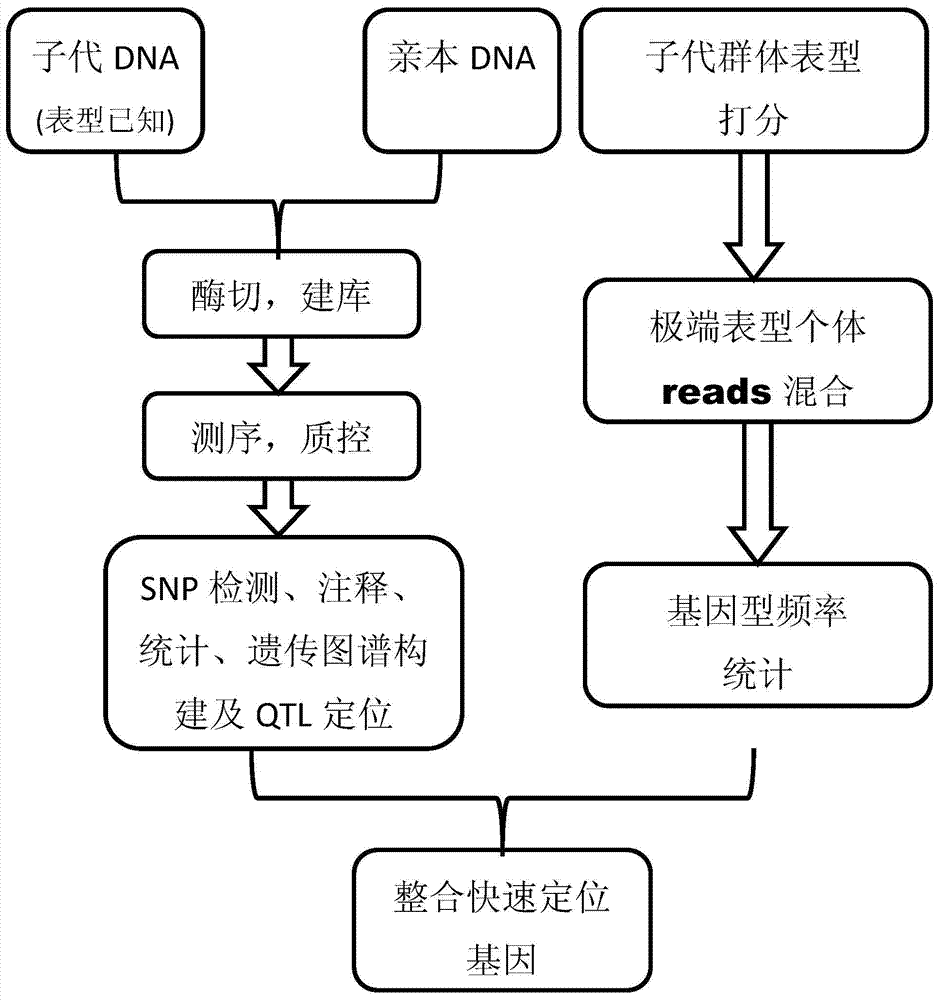
Gene mapping multi-inspection method
ActiveCN104573409AIncrease profitShort cycleMicrobiological testing/measurementSpecial data processing applicationsGene mappingQtl analysis
The invention discloses a gene mapping multi-inspection method. The method is characterized by comprising the steps of 1, sequencing a parent DNA sample and a filial generation DNA sample to obtain a high-precision short segment sequence; 2,comparing the obtained high-precision short segment sequence with a reference sequence, or mutually clustering to obtain accurate SNP information; 3, converting the SNP information into group SNP, further converting into a standard molecular standard format, creating a genetic map, and performing QTL analysis on the basis of the genetic map to obtain QTL seat information; 4, identifying the phenotype information of filial generation DNA sample, accurately counting the phenotype value distribution, grouping the phenotype values according to gradient, respectively blending the high-precision short segment sequences of the filial generation DNA sample, corresponding to the extreme phenotype values, analyzing the genotype frequency distribution, and performing chi-square test and rank sum test to find out related obvious areas; 5, integrating the QTL seat information and the accurate gene mapping information obtained in step 4.
Owner:HANGZHOU HEYI GENE TECH
Molecular marking method for detecting goose hatching egg qualification rate
ActiveCN112430674ABreeding is fast and accurateImprove breeding efficiencyMicrobiological testing/measurementReference genome sequenceAnimal science
The invention relates to the technical field of SNP (Single Nucleotide Polymorphism) molecular markers and provides a molecular marker method for detecting goose hatching egg qualification rate. The method comprises the following steps of comparing whole genome resequencing data to a goose reference genome sequence to obtain whole genome SNP sites, and obtaining seven candidate SNP molecular markers of goose hatching egg qualification rate characters by a whole genome correlation analysis method; and detecting the polymorphism of the seven SNP molecular markers by using a time-of-flight mass spectrometry method and counting the genotype frequency of the seven SNP molecular markers. And one haplotype module is also remarkably related to the egg laying percent of pass. The polymorphism SNP molecular marker and the primer pair comprise at least one of seven SNP molecular markers. According to the universal polymorphic SNP molecular marker for the Sichuan white geese obtained by the screening method, seven effective SNP molecular markers are provided, a database of the SNP molecular markers of the Sichuan white geese is supplemented, individuals with high or low hatching egg percent ofpass can be rapidly detected by utilizing the method, the breeding time and cost of traditional goose breeding are saved, and breeding efficiency is improved; reference is provided for development ofSichuan white goose variety resources and industrial development.
Owner:CHONGQING ACAD OF ANIMAL SCI
Micro-satellite DNA (Deoxyribose Nucleic Acid) method for identifying reproductive mode of termite nest group
InactiveCN102559896AImprove targetingMicrobiological testing/measurementRepetitive SequencesGenotype
The invention relates to a micro-satellite DNA (Deoxyribose Nucleic Acid) method for identifying a reproductive mode of a termite nest group, which comprises the following steps of: collecting above 20 termite samples from a same nest group in the open air, extracting a sample genome DNA and diluting; searching a micro-satellite site related to a termite reproductive mode on GenBank, designing a primer on the side wing of a micro-satellite repetitive sequence by utilizing software Primer Premier 5.0, carrying out electrophoresis on an automatic analyzer after the termite samples DNA are subjected to PCR (Polymerase Chain Reaction) amplification by selecting a micro-satellite primer and determining the genotype of each strip corresponding sample in a electrophoretic diagram by using SAGAGT software; and carrying out chi-square test on observed genotype frequencies and expected genotype frequencies of the termite samples based on a Mendel's law and judging the reproductive mode of the termite nest group. The micro-satellite DNA identification for reproductive modes of ten reticulitermes chinensis nest groups in Changsha by adopting the micro-satellite DNA method shows that the reticulitermes chinensis nest groups include seven simple nest groups, two amplified nest groups and one mixed nest group. The micro-satellite DNA method has the advantages that the operation is simple and convenient, the results are reliable, the reproductive modes of the termite group can be accurately judged, and the important reference value is provided for the formulation of a termite prevention strategy.
Owner:HUAZHONG AGRI UNIV
Genetic marker using MX1 genes as pig production trait and use thereof
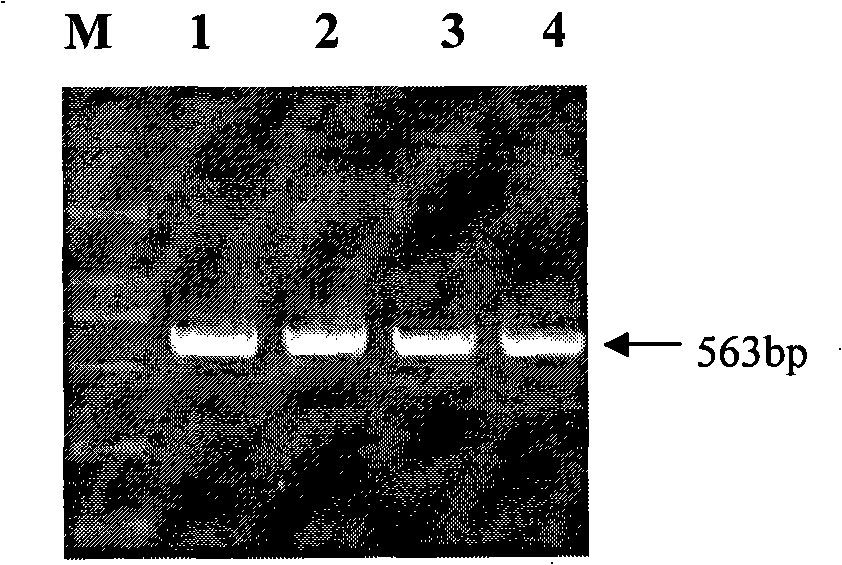
The invention pertains to the technical field of the preparation of a pig genetic marker, more particularly relates to a pig production trait gene MX1 eighth intron mutant site polymorphism detection method and the clone used as the genetic marker of the pig production trait, and the application. Genome DNA is extracted from pig blood, primer PCR amplification is designed, a 563bpDNA sequence is obtained by clone resequencing, and the mutation of A167-G167 is found at the position of 167bp of a sequence list SEQID NO: 1 by sequence comparison analysis, thus causing the polymorphism of HpaII-RFLP. By utilizing the genetic marker, the genotype frequency and the gene frequency of a marker gene and the association analysis of the pig production trait are detected in different swineries, showing that notable association exists between the genetic marker and certain important production trait of the pig. The invention also discloses an SNP typing detection technology of the eighth intron of the pig MX1 gene, thus providing a new marking and detection method for marker auxiliary selection of the pig.
Owner:INST OF ANIMAL SCI & VETERINARY HUBEI ACADEMY OF AGRI SCI
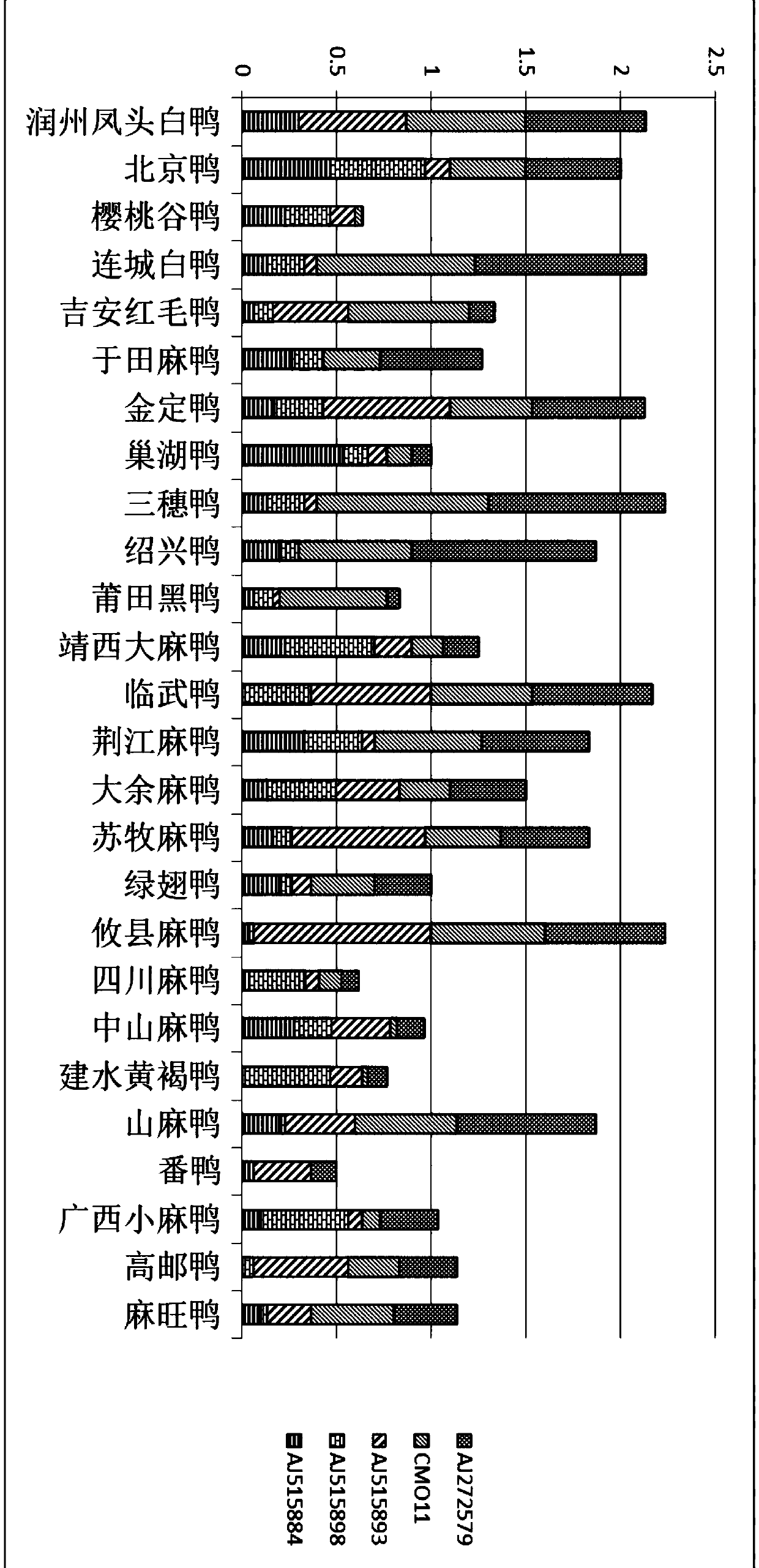
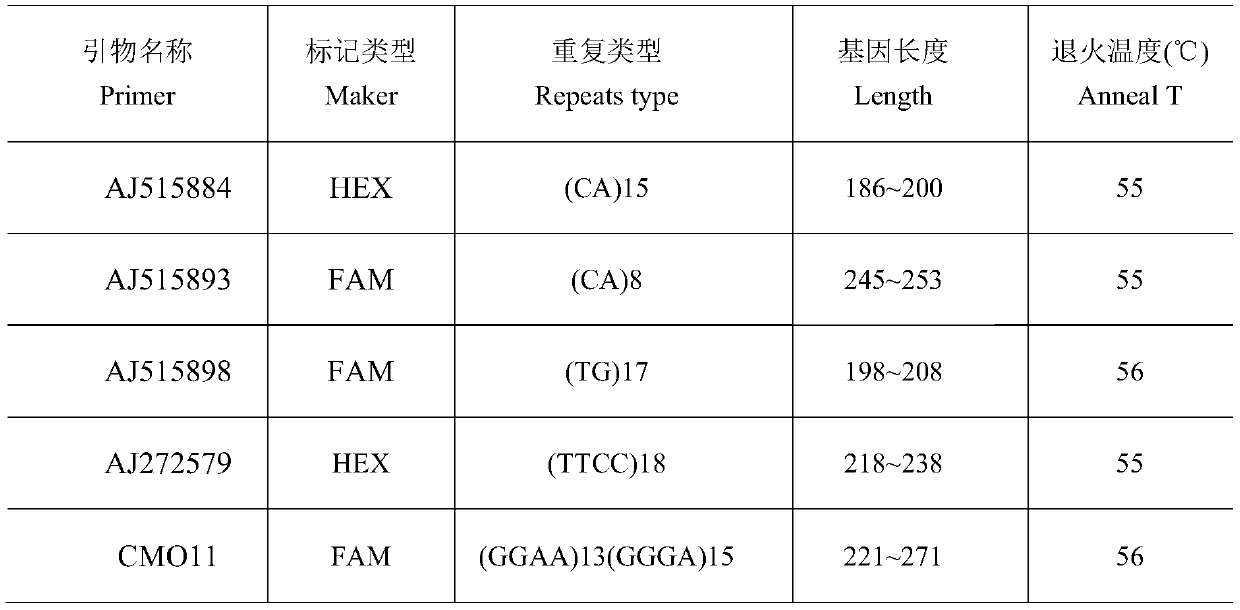
Runzhou crested white duck variety identification method
PendingCN110129454AEfficient use ofEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationStr typingAgricultural science
The invention relates to a Runzhou crested white duck variety identification method, and belongs to the technical field of zootechny and molecular biology. By the aid of microsatellite loci provided by pertinent literature, microsatellite DNA (deoxyribonucleic acid) seats with common genotypes are screened to perform STR (short tandem repeat) type sequencing, data are analyzed by a Microsatellite-Toolkit, genotype frequency is counted, and Runzhou crested white ducks are identified by the aid of common genotype bands of all varieties.
Owner:YANGZHOU UNIV
Genetic variation data-based GDS-Huffman compression method
InactiveCN109192245AEfficient compressionReduce sizeCode conversionSequence analysisProgramming languageGenotype
The invention relates to a genetic variation data-based GDS-Huffman compression method. Genotypes in a GVCF file are coded with Huffman codes according to a genotype frequency on the basis of a GDS compression method; and integer type fields in the GVCF file are coded with a length-varying integer type coding mode; and therefore, a compressed GDS file can be obtained.
Owner:SUN YAT SEN UNIV
Method and system used for deducing Han, Tibetan or Wei population source of individual with unknown source
ActiveCN103146820BEasy to classifyImprove accuracyMicrobiological testing/measurementGeneticsGenotype
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
Multiple testing methods for gene mapping
ActiveCN104573409BIncrease profitShort cycleMicrobiological testing/measurementSpecial data processing applicationsGene mappingSequence analysis
The invention discloses a multiple detection method for gene positioning, which is characterized in that it comprises: step 1: sequence the DNA samples of parents and offspring to obtain high-precision short fragment sequences; step 2: sequence the obtained high-precision DNA samples The short fragment sequence is compared with the reference sequence or clustered with each other to obtain accurate SNP information; Step 3: convert the SNP information into a population SNP, and further convert it into a standard molecular standard format to construct a genetic map, and on the basis of the genetic map Step 4: Identify the phenotypic information of the progeny DNA samples, and accurately count the distribution of phenotypic values, group the phenotypic values according to gradients, and group the extreme phenotypic values corresponding to the offspring The high-precision short fragment sequences of the first-generation DNA samples are mixed separately, the genotype frequency distribution is analyzed, and the relevant significant regions are found through the chi-square test and the rank sum test; Step 5: Integrate the QTL locus information and Step 4 to obtain accurate gene positioning information.
Owner:HANGZHOU HEYI GENE TECH
A method for establishing an animal paternity system based on genome-wide str
ActiveCN104480205BShort method cycleIncrease screening throughputMicrobiological testing/measurementAgricultural scienceGermplasm
Owner:BGI FORENSIC TECH (SHENZHEN) CO LTD
A method for individual selection and optimization of chicken conservation population
ActiveCN106086172BReduce the level of inbreedingSolve the problem that individual fine matching cannot be carried outMicrobiological testing/measurementGenomic sequencingGenetic diversity
The invention discloses a chicken preserved population individual apolegamy optimizing method. Blood DNA of chicken preserved population breed reserving cock and hen individuals is extracted, sequencing is carried out through a reduced genomic sequencing technology, and the sequence is compared with a sequence of the GenBank database reference chicken breed to find SNP loci existing in different individuals. The individual own inbreeding coefficient and the interindividual phylogenetic relationship coefficient are calculated through gene frequency and genotype frequency of the SNP loci, individual apolegamy is optimized according to the statistics value, mating combination of individuals with an overlarge inbreeding coefficient and individuals with an overlarge within-family phylogenetic relationship coefficient is avoided, and the purposes of reducing the preserved population inbreeding level and maintaining the genetic diversity total quantity steady state are achieved. By means of the method, the problem that fine individual apolegamy cannot be carried out depending on genealogy information in the chicken breed resource protection process can be effectively solved, the preserved population inbreeding level can be effectively reduced, the genetic diversity steady state is maintained, and the breed preservation effect and efficiency are improved.
Owner:JIANGSU INST OF POULTRY SCI
Disease susceptibility risk prediction device
ActiveCN105069322BHelp with health managementTo achieve the effect of "preventive cure"Special data processing applicationsDisease riskArray data structure
The invention relates to bioinformatics and provides a disease susceptibility risk prediction method and device. The disease susceptibility risk prediction method comprises the steps: providing a database including disease morbidity data, SNP locus genotype frequency data, and risk allele homozygous and heterozygous genotype OR value data aiming at each disease-associated SNP locus; receiving information of a to-be-predicted individual; calculating to obtain a comprehensive disease susceptibility risk array of diseases interested by the to-be-predicted individual; and generating a comprehensive disease susceptibility risk dynamic change curve of the individual in an appointed age range. According to the disease susceptibility risk prediction method and device, two factors , including individual inheritance and environment factors, are considered at the same time when the disease susceptibility risk of an individual is calculated, so that calculating results greatly conform to objective reality; according to the obtained disease susceptibility risk age change curve of the individual, the individual not only can learn more precise immediate disease susceptibility risk, but also can continuously understand the susceptibility change tendency of various diseases along with the growth of age.
Owner:上海尔云信息科技有限公司
Genetic marker using MX1 genes as pig production trait and use thereof
The invention pertains to the technical field of the preparation of a pig genetic marker, more particularly relates to a pig production trait gene MX1 eighth intron mutant site polymorphism detection method and the clone used as the genetic marker of the pig production trait, and the application. Genome DNA is extracted from pig blood, primer PCR amplification is designed, a 563bpDNA sequence is obtained by clone resequencing, and the mutation of A167-G167 is found at the position of 167bp of a sequence list SEQID NO: 1 by sequence comparison analysis, thus causing the polymorphism of HpaII-RFLP. By utilizing the genetic marker, the genotype frequency and the gene frequency of a marker gene and the association analysis of the pig production trait are detected in different swineries, showing that notable association exists between the genetic marker and certain important production trait of the pig. The invention also discloses an SNP typing detection technology of the eighth intron ofthe pig MX1 gene, thus providing a new marking and detection method for marker auxiliary selection of the pig.
Owner:INST OF ANIMAL SCI & VETERINARY HUBEI ACADEMY OF AGRI SCI
A SNP site related to growth traits of Chlamys farreri and its application
ActiveCN105112424BImprove growth traitsAvoid crossbreedingMicrobiological testing/measurementFermentationNucleotideNucleotide sequencing
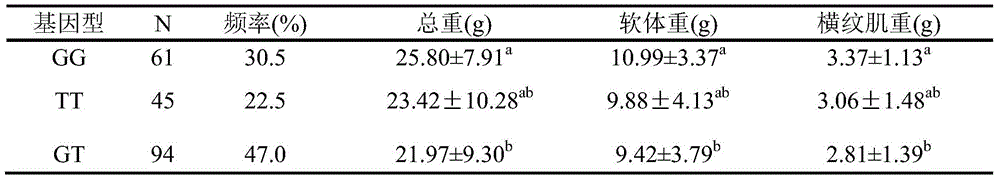
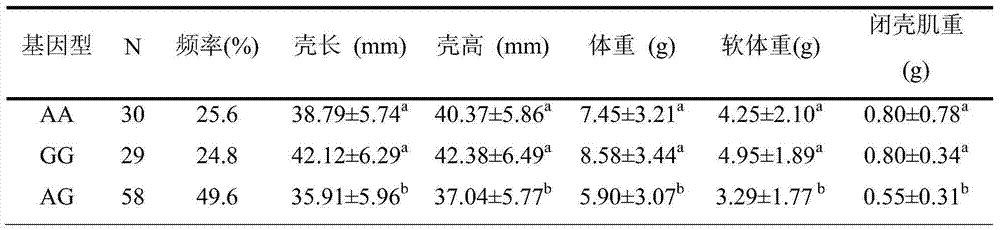
The invention provides an SNP (single nucleotide polymorphism) site relevant to chlamys farreri growth traits. The site is the 846th bit of Smad gene with the nucleotide sequence being SEQ ID NO:2, and the basic group is G or T. The invention also provides a serotype specific primer and a probe for detecting the SNP site. A high-resolution melting curve technology is used for detecting the site polymorphism in a chlamys farreri group, and analyzing the relevance between the site genotype frequency and the farreri production traits. The site C.846G is greater than T and is significantly relevant to the important growth traits such as chlamys farreri weight, soft weight and closed shell muscle weight; the growth traits of GG type individuals are significantly higher than those of GT type individuals (P is smaller than 0.05).
Owner:OCEAN UNIV OF CHINA
Reagent kit for detecting diabetes
ActiveCN109554466ARapid identificationMicrobiological testing/measurementDNA/RNA fragmentationNucleotide sequencingBiology
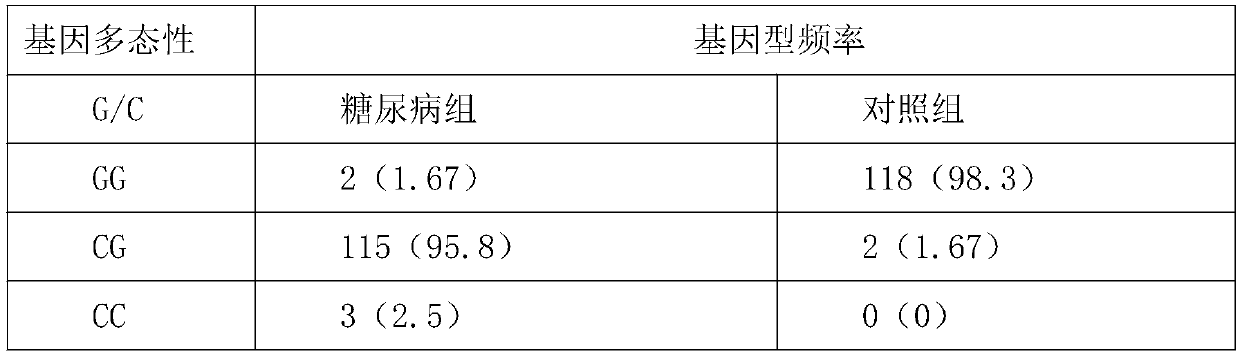
The invention provides a reagent kit for detecting diabetes. The reagent kit contains an SNP site relevant to properties of type II diabetes, the site is the 166th of PDP1 of which the nucleotide sequence is SEQID NO:1, and a basic group is G or C. The invention also provides a probe and typing primer for detecting the SNP site. The site polymorphism is detected in human diabetes potential groupsthrough a high-resolution melting curve technique, and relevance of the genotype frequency of the site and the properties of the diabetes is analyzed. The site can be used as a fast detection index tobe used for large-scale potential patient screening.
Owner:广东创晟控股集团有限公司
A kind of primer, probe and method for detecting drug resistance gene mutation of Aedes albopictus
ActiveCN109234408BMicrobiological testing/measurementDNA/RNA fragmentationGenes mutationInsecticide resistance
The invention provides a primer, a probe and a method for detecting the drug resistance gene mutation of Aedes albopictus. The primers, TaqMan-MGB probe and detection method provided by the present invention can realize fast, simple, sensitive and accurate monitoring of kdr allele type, genotype frequency, etc., and can quickly and accurately grasp Aedes albopictus and pyrethroid Status of KDR alleles associated with ester insecticide resistance to understand the status of resistance genes in the genetic structure of populations. Therefore, the primers, TaqMan-MGB probe, kit and detection method provided by the present invention are a new product and method that can accurately and conveniently predict the occurrence and development of drug resistance in Aedes albopictus.
Owner:宁波国际旅行卫生保健中心
Rapid positioning method for industrial hemp trait related genes
PendingCN112575104ARapid positioningMicrobiological testing/measurementProteomicsNucleotideCorrelation analysis
The invention discloses a rapid positioning method for industrial hemp trait related genes, and belongs to the technical field of industrial hemp research. The method comprises the following steps: variety selection, extreme group construction, specific site amplified fragment library construction and high-throughput sequencing, marker development, correlation analysis, gene annotation and real-time fluorescent quantitative PCR. The method comprises the following steps: constructing a high-low mixing pool for F1-generation groups obtained by hybridizing parents with relatively large target character differences, re-sequencing the parents, sequencing the high-low mixing pool by adopting a simplified genome deep sequencing technology, developing an SLAF tag, carrying out single nucleotide polymorphism detection, and carrying out marker correlation analysis on genotype frequency differences among the mixing pools. According to the method, a target character related candidate region is obtained, most plant populations at least need to be propagated to F2 generations to perform gene localization, and due to the high heterozygosity characteristic of industrial hemp, gene localization isrealized through F1 generations, so that rapid localization of industrial hemp genes can be realized in a high-efficiency and low-cost manner.
Owner:DAQING BRANCH OF HEILONGJIANG ACAD OF SCI
Hu sheep PLIN2 gene SNP detection method and application of Hu sheep PLIN2 gene SNP detection method in meat quality character early screening
PendingCN114854878ASpeed up breedingMicrobiological testing/measurementDNA/RNA fragmentationMarker-assisted selectionMedicine
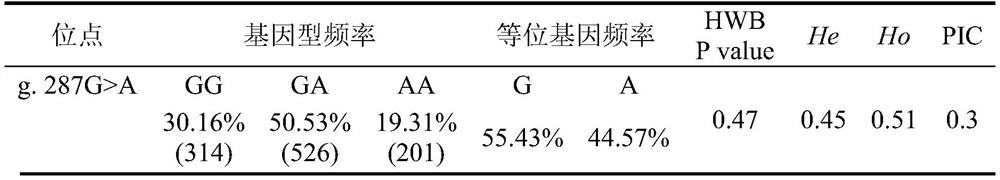
The invention discloses a Hu sheep PLIN2 gene SNP (Single Nucleotide Polymorphism) detection method and application of the Hu sheep PLIN2 gene SNP detection method in early screening of meat quality characters. According to the invention, Hu sheep tissue DNA is extracted, an iMLDR technology is used for typing, allele frequency and genotype frequency are calculated, and Hu sheep back longissimus fatty acid percentage and PLIN2 gene mutation site g.287Ggt; correlation analysis is carried out on the site A, the polymorphism of the site and the existing molecular marker obviously related to the percentage of beneficial fatty acid are disclosed, and a basis is provided for selecting and breeding a new sheep strain with excellent meat quality through marker-assisted selection.
Owner:甘肃润牧生物工程有限责任公司
SNP locus related to growth characteristics of patinopecten yessoensis and detection and application thereof
InactiveCN103740729BHigh trait valueAvoid crossbreedingMicrobiological testing/measurementClimate change adaptationInsulin-like growth factor bindingNucleotide sequencing
The invention provides an SNP locus related to growth characteristics of patinopecten yessoensis. The locus is the 1054 site of IGFBP (Insulin Like Growth Factor Binding Proteins) genes of which the nucleotide sequence is shown as SEQ ID NO:2 and has the base of A or G. The invention also provides a probe for detecting the SNP locus and a parting primer. The transcription sequence of the IGFBP genes in the patinopecten yessoensis is subjected to sequencing and Clustal comparison, and three SNP loci are screened. The site polymorphism is detected in the patinopecten yessoensis group by using a high resolution melting curve technology, and correlation between the site genotype frequency and the growth characteristics of patinopecten yessoensis is analyzed. The locus C.1054A>G is obviously related to important growth characteristics such as the height, shell length, weight, soft weight and adductor muscle weight of the patinopecten yessoensis, the growth characteristics of the AG type individuals are obviously lower than those of AA and GG type individuals (P is less than 0.05), and the characteristic value of the GG type is the highest. Therefore, individuals of which the genotype is GG on the locus can be preferentially selected during production and are used as parents for performing high-yield patinopecten yessoensis breeding or performing large-scale breeding.
Owner:OCEAN UNIV OF CHINA
SNP (single nucleotide polymorphism) site relevant to chlamys farreri growth traits and application of SNP site
ActiveCN105112424AHigh productivity traitsImprove growth traitsMicrobiological testing/measurementFermentationBiotechnologyNucleotide
The invention provides an SNP (single nucleotide polymorphism) site relevant to chlamys farreri growth traits. The site is the 846th bit of Smad gene with the nucleotide sequence being SEQ ID NO:2, and the basic group is G or T. The invention also provides a serotype specific primer and a probe for detecting the SNP site. A high-resolution melting curve technology is used for detecting the site polymorphism in a chlamys farreri group, and analyzing the relevance between the site genotype frequency and the farreri production traits. The site C.846G is greater than T and is significantly relevant to the important growth traits such as chlamys farreri weight, soft weight and closed shell muscle weight; the growth traits of GG type individuals are significantly higher than those of GT type individuals (P is smaller than 0.05).
Owner:OCEAN UNIV OF CHINA
Application of single nucleotide polymorphism of goat MMP9 gene in early selection of lactation character
ActiveCN113913531ASpeed up buildSpeed up breedingFood processingMicrobiological testing/measurementAnimal scienceMedicine
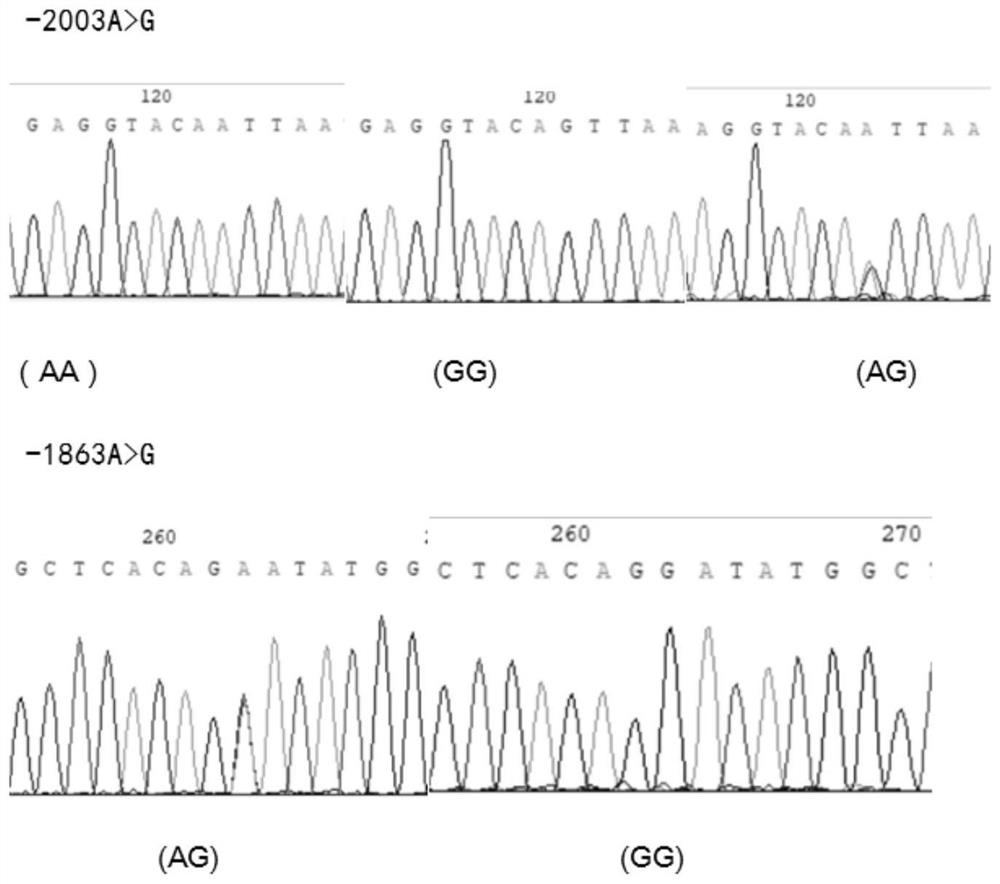
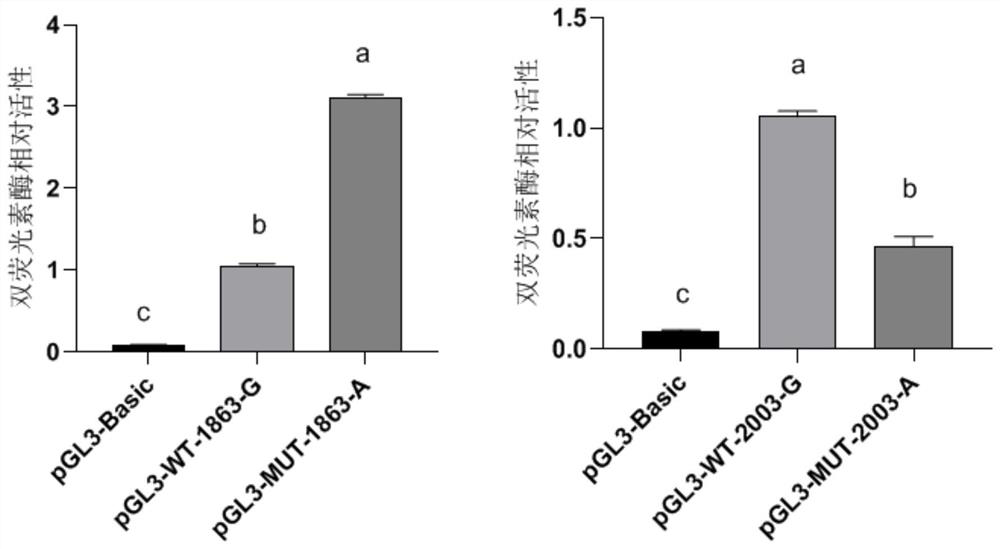
The invention discloses an application of single nucleotide polymorphism of a goat MMP9 gene in early selection of lactation traits. Two new SNP loci (c.-2003A > G and c.-1863A > G) located in a 5'regulation region of the MMP9 gene of the milk goat are identified for the first time, gene frequency and genotype frequency analysis and correlation analysis with milk goat lactation traits are carried out, and the result shows that the SNP loci can be used as marker loci of the milk goat lactation traits, so that the marker loci can be used as marker loci of the milk goat lactation traits. The method can be used for accelerating establishment of goat populations with excellent lactation characters, so that the breeding speed of improved varieties is increased.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
A method and application for detecting single nucleotide polymorphism of sheep spata6 gene
ActiveCN110241227BPromote amplificationEasy to unlinkMicrobiological testing/measurementProteomicsNucleotideMedicine
The invention discloses a method for detecting the single nucleoside polymorphism of sheep SPATA6 gene, comprising the following steps: S1 sample selection, S2 cDNA pool construction, S3 PCR amplification, S4 product sequencing and single nucleotide polymorphism analysis Type, S5 sheep SPATA6 gene single nucleotide polymorphism site frequency statistics and its correlation analysis with testis phenotype; Genotype frequency calculation, typing results of fluorescent multiple enzyme ligation reaction technology, testicular phenotype data, and SAS (9.2) software analysis data, and then fully constructed the 72272th T>C mutation of testicular phenotype and spermatogenesis-related gene 6 Relationship of single nucleotide polymorphisms to provide selection markers for breeding high fecundity sheep.
Owner:甘肃润牧生物工程有限责任公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com