Genetic variation data-based GDS-Huffman compression method
A compression method and gene mutation technology, applied in the field of life omics analysis, can solve the problem of not considering the excessive size of the intermediate file, and achieve the effect of solving large file processing, efficient compression, and reducing size
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
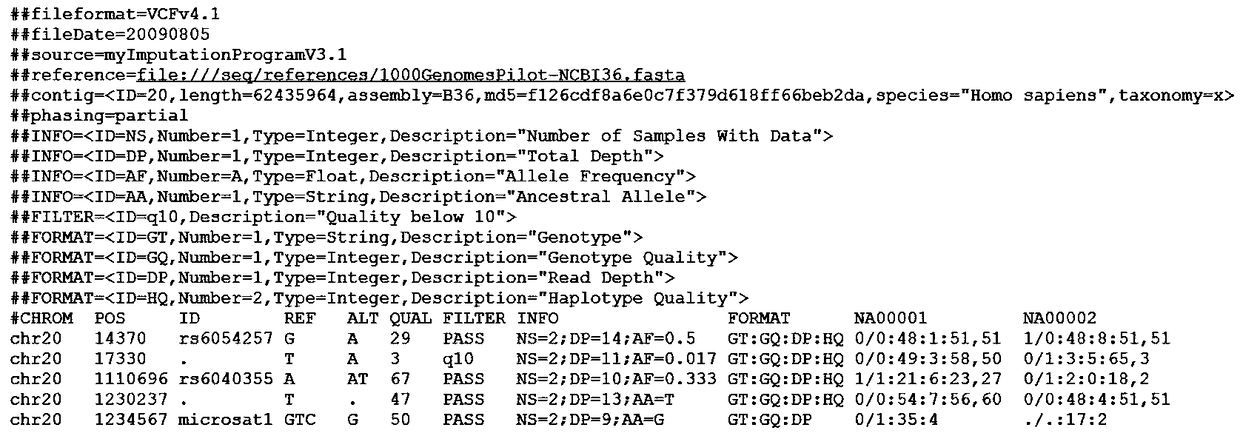
[0027] In this embodiment, for the GVCF file, based on the GDS compression method, the genotype frequency characteristics are utilized, and the genotype is encoded by Huffman coding, so as to compress the GVCF file more efficiently.
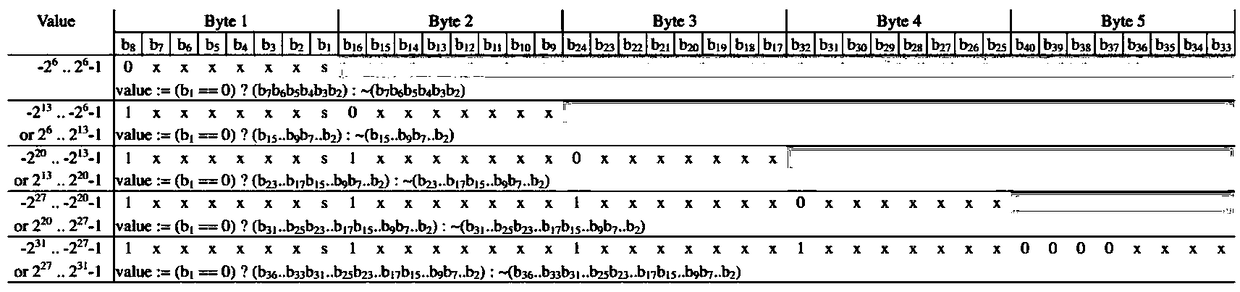
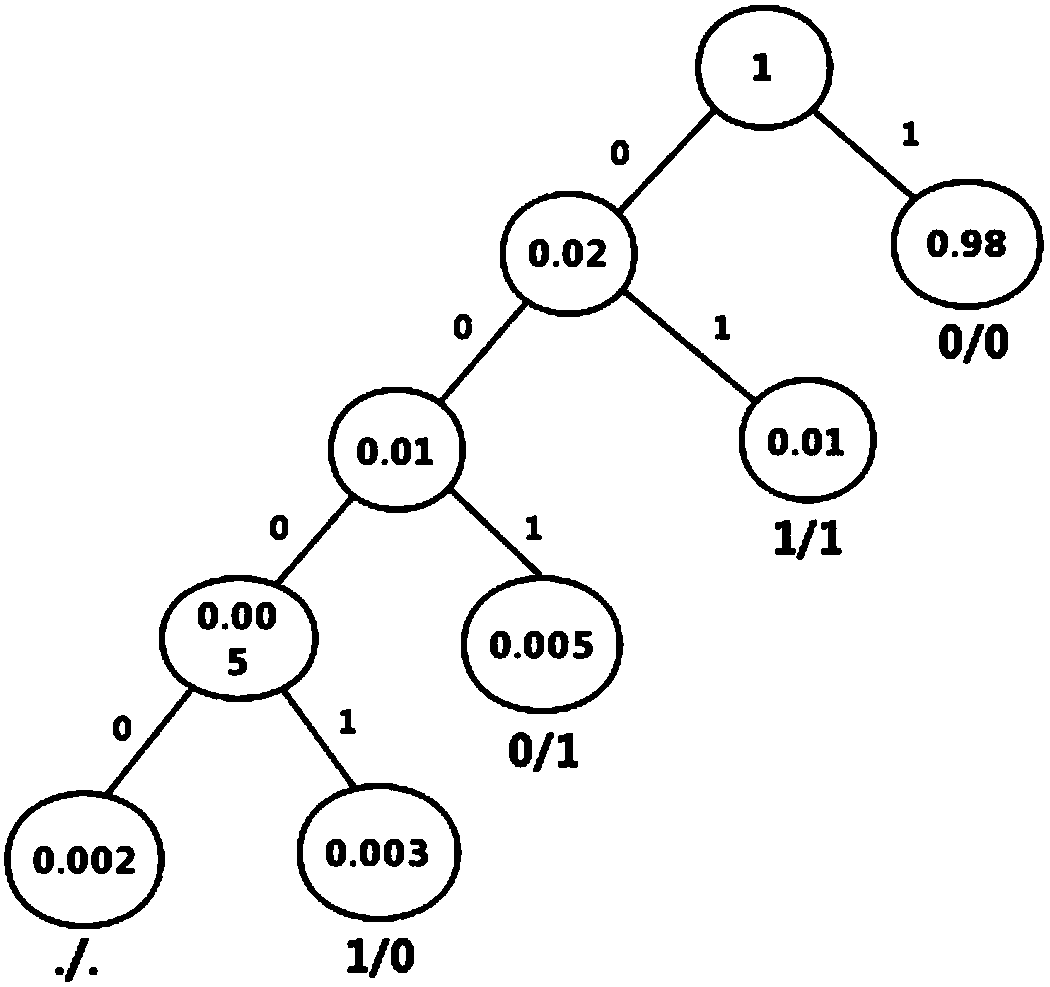
[0028] The Huffman coding tree constructed according to the genotype frequency is as follows image 3 shown. The coding is shown in Table 2. Genotypes will be coded according to the coding table. The integer field in the GVCF file is encoded in a variable-length integer encoding manner.
[0029] Table 2 Genotype Huffman code
[0030] genotype
coding
0 / 0
1
1 / 1
01
0 / 1
001
. / .
0000
1 / 0
0001
[0031] Use the above method for Figure 4 The mutation information in is encoded, and the result is:
[0032] GT:100101
[0033] GQ:00010100 00010100 00010100
[0034] DP: 01000000 01100000 10000000 00000001
[0035] In this embodiment, the GDS-Huffman compression method is applied in the w...
Embodiment 2
[0038] Table 3 is a test of the compression performance of the GDS-Huffman compression method. This test is aimed at the GVCF file. The original data comes from the whole exome sequencing data in the 1000 Genomes Project (The 1000 Genomes Project). The detailed information of the samples can be queried on the official website of the 1000 Genomes. These data have been compared to the reference genome and passed CRAM format files for compression by CRAM. Firstly, the CRAM file was preprocessed by Samtools to obtain the FastQ format file of the original sequence, and then these samples were processed according to the whole exome analysis process to obtain the GVCF file of each sample. Take a GVCF file as input and compress it using the GDS-Huffman compression method. The average compression rate reaches 5.1%, and the compression rate is 4.1M / s.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com