SNP locus related to growth characteristics of patinopecten yessoensis and detection and application thereof
A technology for growth traits and scallops, applied in the field of molecular breeding genetic marker screening, can solve the problems of unseen IGF system gene research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] The cDNA sequence cloning of the scallop IGFBP gene in the present invention comprises the following steps:
[0023] a) extraction of total RNA from adductor muscle of scallop scallop;
[0024] b) synthesis of cDNA first strand;
[0025] c) Obtaining the full-length cDNA sequence of the target gene;
[0026] d) Bioinformatics analysis of the target gene.
[0027] The specific operation is as follows:
[0028] a) Extraction of total RNA from Ezo scallop. Total RNA was extracted from adductor muscle of Ezo scallop according to the Trizol method.
[0029] b) Synthesis of cDNA first strand. Using 2μg total RNA as a template, add 1μl 20μM Oligo d(T) 18 , RNase&DNase-free H 2 O to make up to 13μl, mix and centrifuge. Incubate at 70°C for 10 min, and immediately place on ice to open the RNA secondary structure. Add in sequence: 5μl 5×M-MLV Buffer, 5μl dNTP (2.5mM), 1μl RNasin (40U / μl), 1μl M-MLV (200U / μl); mix well and centrifuge at 42°C for 90min. 94°C, 5min, to ina...
Embodiment 2
[0035] Example 2: Screening analysis of SNP sites related to growth traits in the IGFBP gene of Ezo scallop and its method for assisting high-yielding scallop breeding
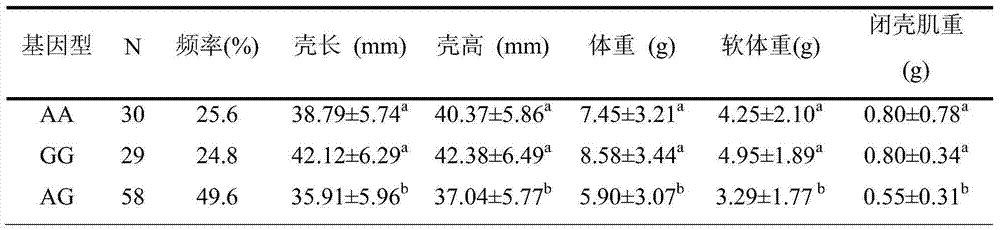
[0036] The correlation analysis between the growth traits-related SNP site C.1054A>G and the growth traits in the IGFBP gene of the scallop scallop and the method for its application in the breeding of the high-yielding scallop include the following steps:
[0037] a) extraction of scallop genome DNA;
[0038] b) SNP site screening of the IGFBP gene of the scallop;
[0039] c) Design of primers and probes for locus C.1054A>G typing;
[0040] d) SNP C.1054A>G typing;
[0041] e) Correlation analysis between C.1054A>G genotype and growth traits;
[0042] f) The method of SNP C.1054A>G assisted breeding of high-yielding Chlamys farreri.
[0043] The specific operation is as follows:
[0044] a) Extraction of DNA from Ezo scallop. Take about 0.1 g of adductor muscle, add 500 μl of STE lysis buffer, cut it into...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com