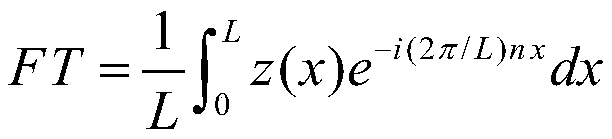A method for predicting protein self-interactions
A predictive method, protein technology, applied in the field of machine learning and bioinformatics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0051] Below in conjunction with accompanying drawing, preferred embodiment of the present invention is described further:
[0052] Such as figure 1 As shown, a method for predicting protein self-interactions, including the following steps:
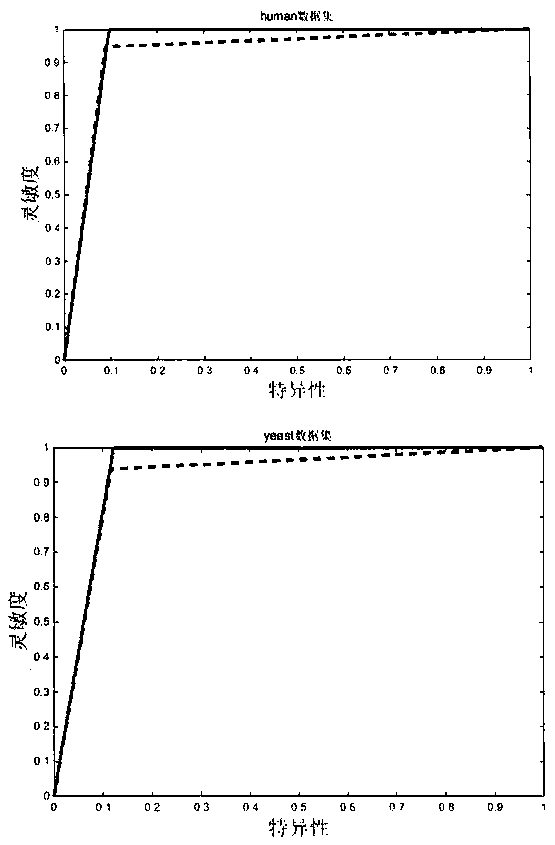
[0053] Step 101: selection and establishment of data sets, using the two gold standard data sets of human and yeast in the UniProt database to construct a data set for predicting protein self-interaction;
[0054] Step 102: Generation of PSSM matrix, the position of each protein sequence can be expressed as a matrix of M × 20, wherein M represents the number of residues in a protein, and the columns of the matrix represent 20 amino acids. By using BLAST Position-specific (PSI-BLAST) converts each protein into a PSSM matrix;
[0055] Step 103: Fourier descriptor extracts eigenvalues. First, multiply the PSSM transposition matrix of each protein transformation with the original PSSM matrix, so that each protein sequence is converted into ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com