Three-dimensional substructure-based drug molecule comparison method
A three-dimensional substructure, drug molecule technology, applied in molecular design, chemical structure search, special data processing applications, etc., can solve the need for 3D substructure retrieval and comparison without considering the importance of substructure parts to biological activity and other issues to achieve the effect of improving flexibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1: Comparison of comprehensive similarity between two molecules when only one query substructure is defined
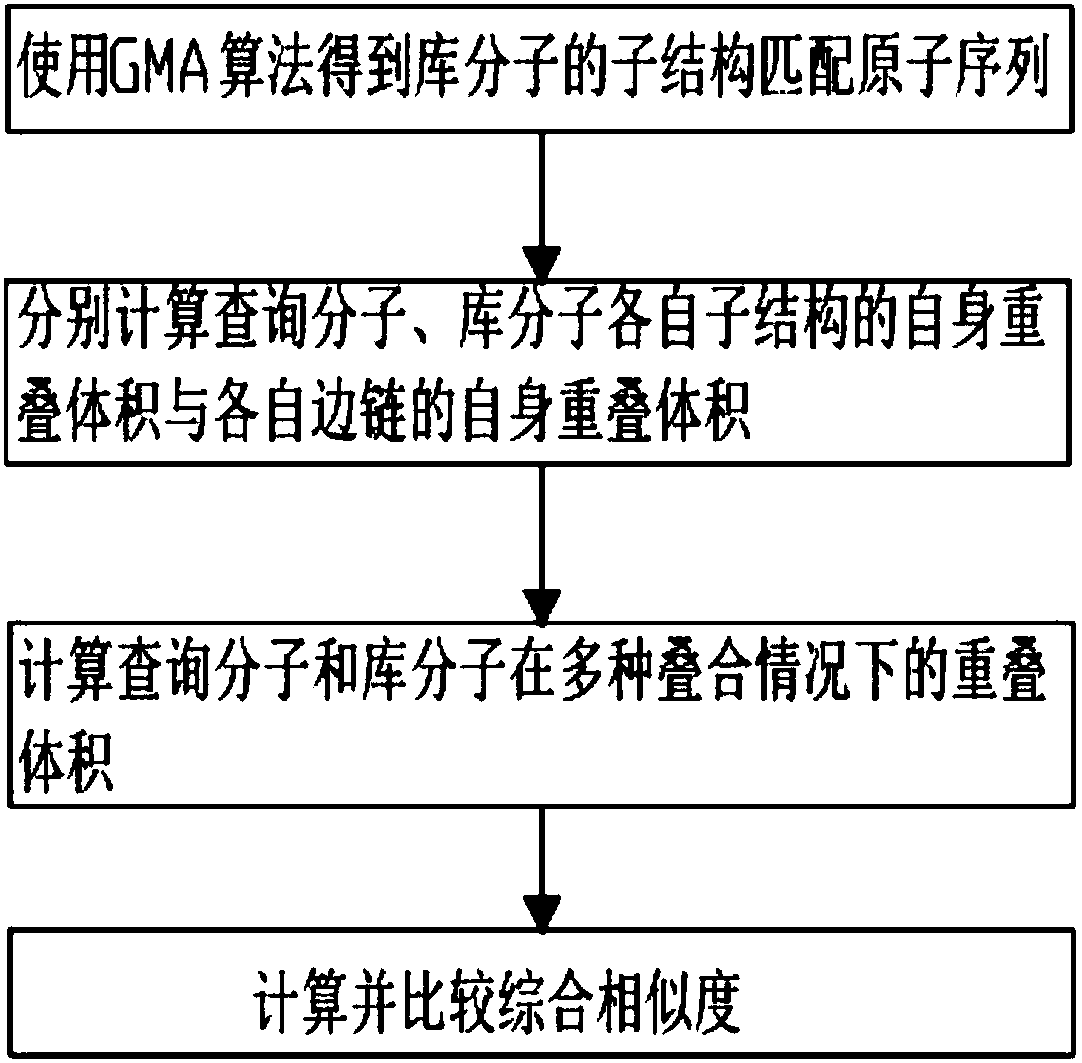
[0041] Such as figure 1 Shown, method of the present invention comprises the steps:
[0042] Step (1), read the two-dimensional or three-dimensional structure information of the library molecule, define the query substructure, and obtain the substructure matching atomic sequence of the library molecule, specifically including:
[0043] The substructure defined in the query molecule is a query substructure, and the defined substructure can be one or more, and the atomic sequence of the query substructure is obtained;
[0044] Read in the structural information of the query substructure, the structure information including the type of each atom and bond in the query substructure;
[0045] reading in two-dimensional or three-dimensional structural information of library molecules, said two-dimensional or three-dimensional structural information including ...
Embodiment 2
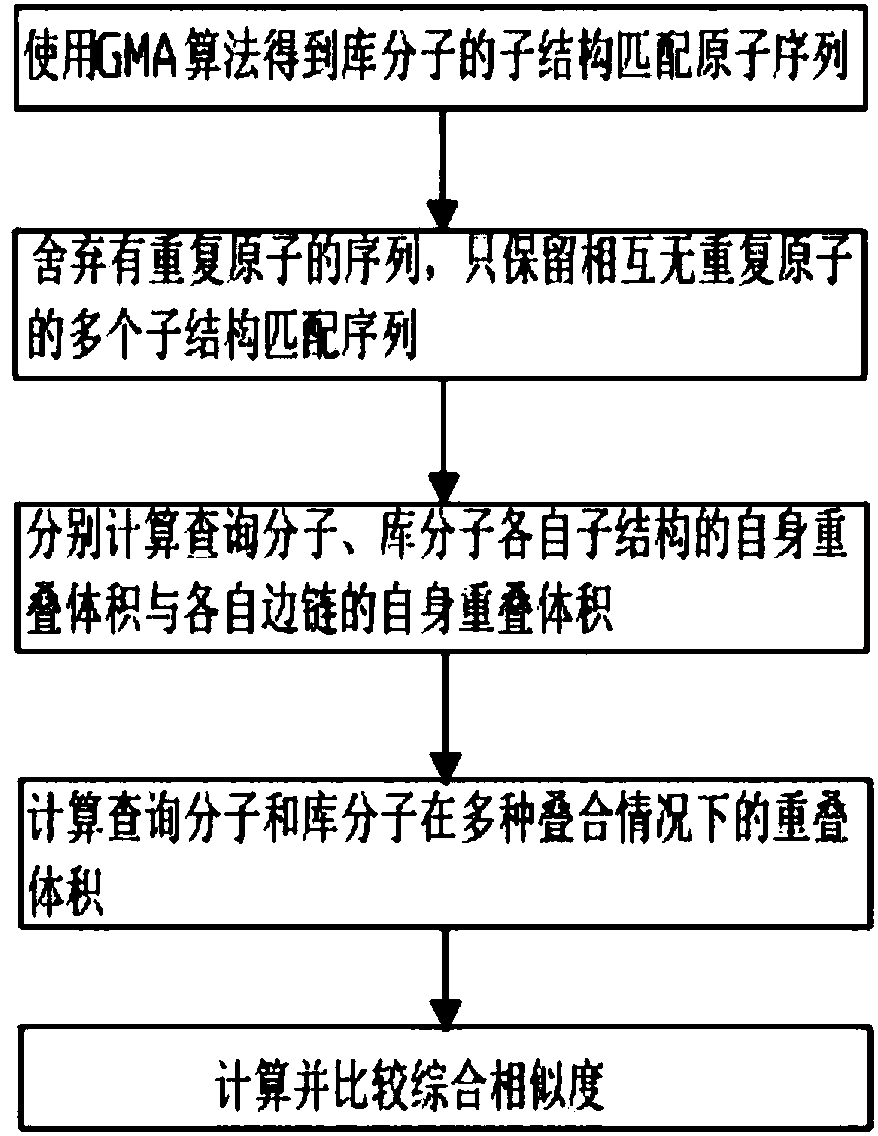
[0064] Embodiment 2: The comparison of the comprehensive similarity between two molecules when multiple query substructures are defined, such as image 3 shown.
[0065] When multiple query substructures are defined, multiple query substructures cannot overlap when defined. After using the GMA algorithm to obtain the substructure matching atomic sequence of the library molecule, judge whether there are repeated atoms in the multiple substructure matching atomic sequences of the library molecule, discard the sequence with repeated atoms, and only keep the multiple substructure matching sequences without repeated atoms; Multiple substructure matching sequences without repeating atoms are merged into one substructure sequence, which is regarded as a substructure, and all matching substructure matching atomic sequences of the library molecules are found by the method of Example 1, and the self of each substructure of the two molecules is calculated The overlapping volume is compa...
Embodiment 3
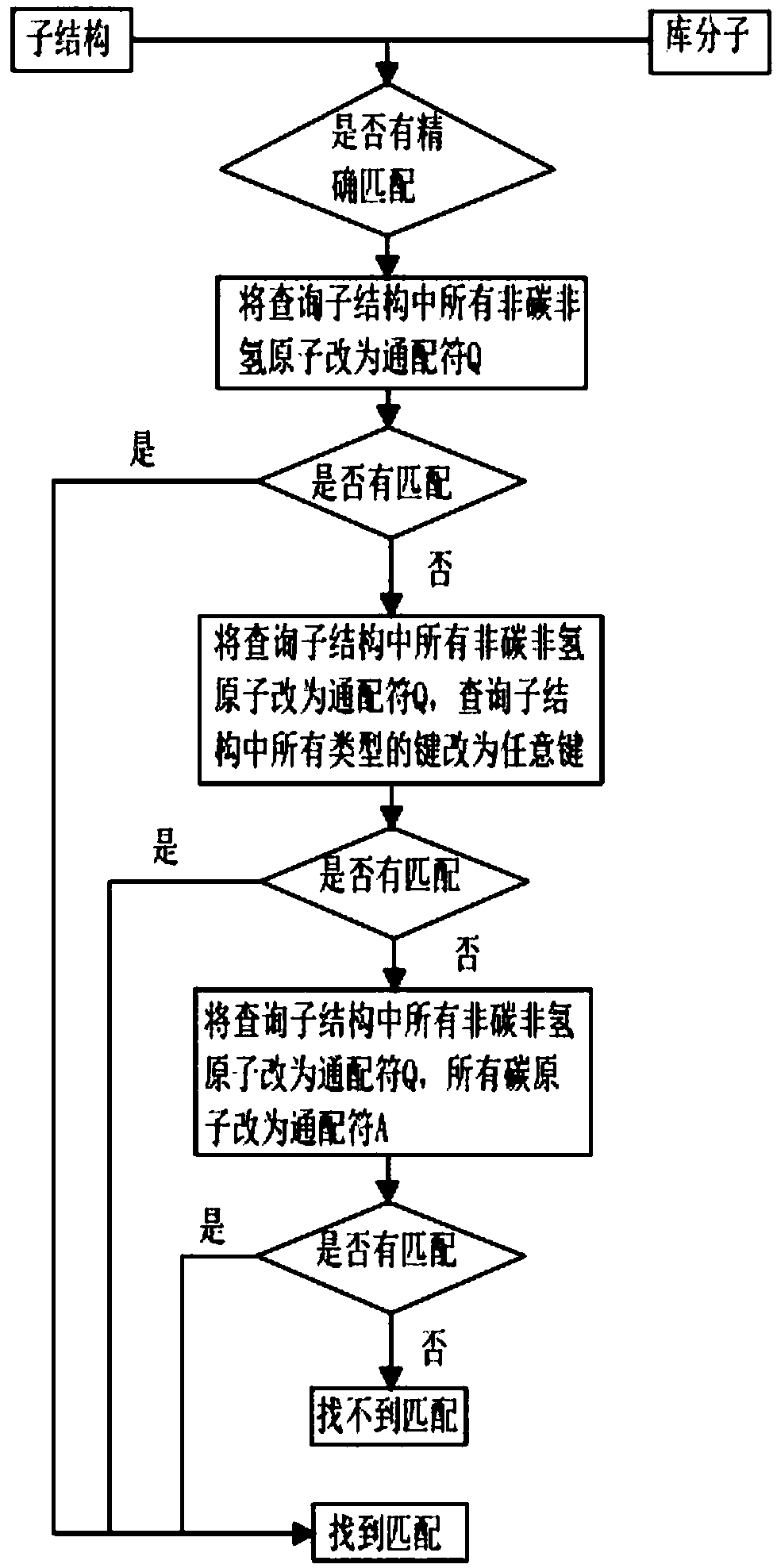
[0066] Embodiment 3: The comparison of molecular comprehensive similarity when defining a query substructure and one or more exogenous substructures, such as Figure 4 shown.
[0067] When the defined query substructure is one, one or more exogenous substructures can be defined (exogenous substructures are substructures that are not in the query molecule but have similar functions to the query substructure), using the GMA algorithm After obtaining the matching atomic sequence of the substructure corresponding to the query substructure or the foreign substructure of the library molecule, there is no need to judge whether there are repeated atoms in all the matching atomic sequences. The exogenous substructure will be regarded as a separate substructure, continue to match the substructure of the library molecule using the method of Example 1, use the method of Example 1 to find all matching substructure matching atomic sequences of the library molecule, and calculate the respect...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com