Method for identifying unconventional cells in pathological section
A technology of pathological slices and identification methods, applied in the field of identification of unconventional cells in pathological slices, can solve the problems of increased error rate, heavy work, time-consuming, etc., and achieve effective classification and good test results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0068] In order to further understand the present invention, the method for identifying unconventional cells in a pathological section provided by the present invention will be specifically described below in conjunction with specific implementation methods, but the present invention is not limited thereto. The non-essential improvements and adjustments mentioned above still belong to the protection scope of the present invention.
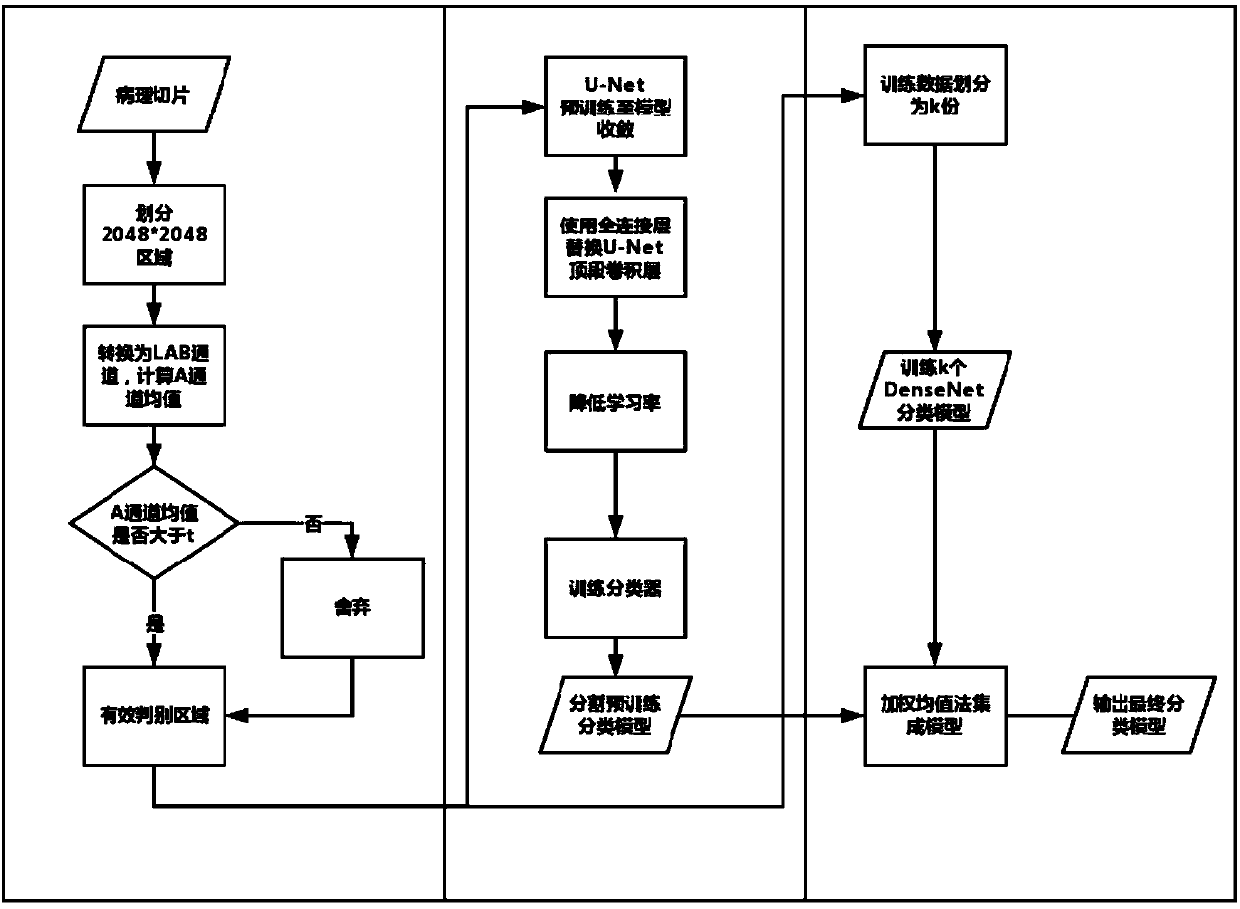
[0069] A method for identifying unconventional cells on pathological slices, the specific steps are:
[0070] 1) Pathological slice preprocessing and valid area discrimination
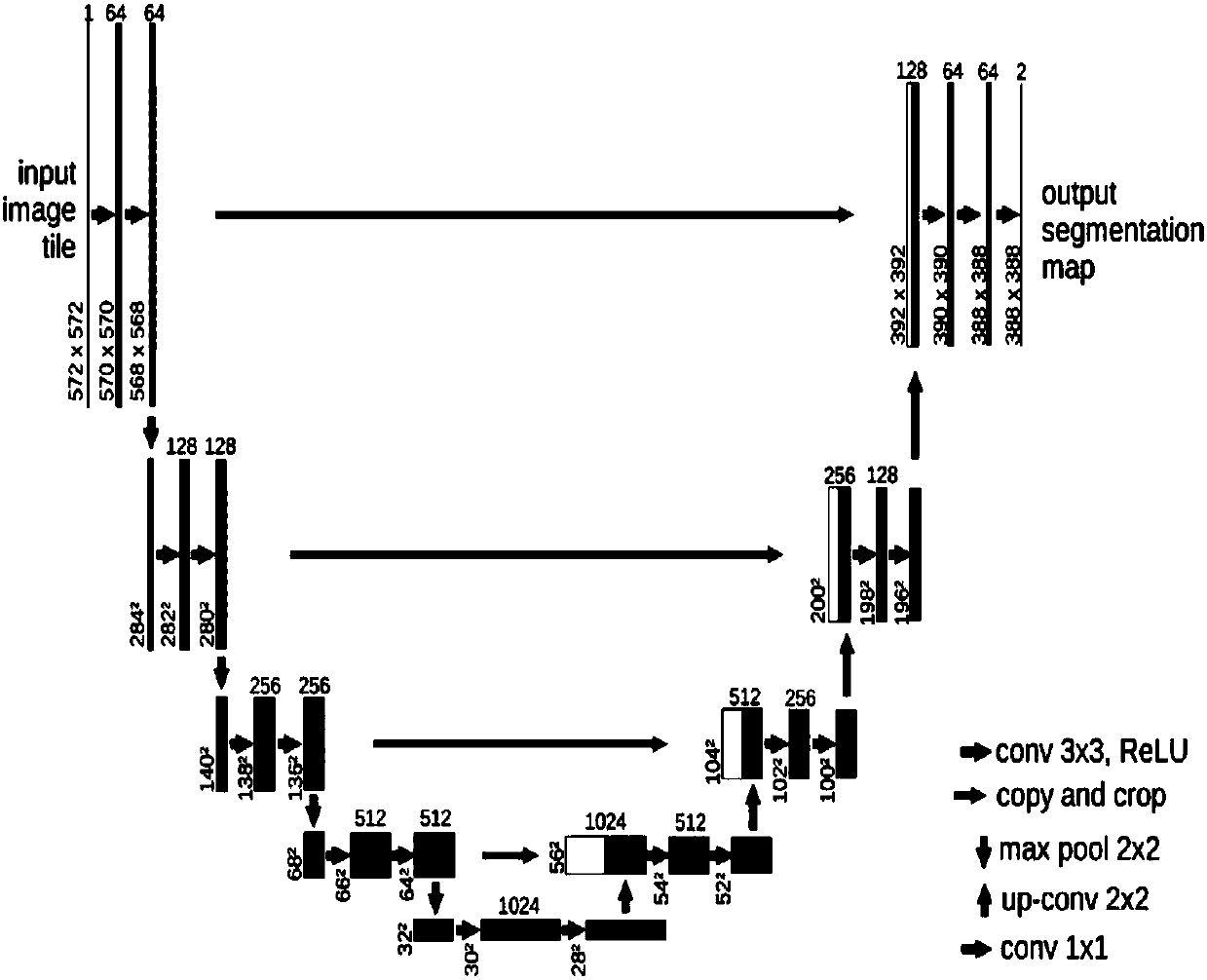
[0071] In the present invention, the input data is 20x enlarged pathological slices, which are divided into areas with a pixel resolution of 2048*2048 and stored separately.
[0072] Convert the region with a pixel resolution of 2048*2048 above into LAB channels, and use the region where the average value of channel A exceeds the threshold t=132 as an effective discriminat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com