Fluorescent-labeled 32-plex InDels composite amplification system and application thereof
A compound amplification system and fluorescent labeling technology, applied in the field of compound amplification system and fluorescently labeled 32-plexInDels compound amplification system, can solve the problem of low recognition ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
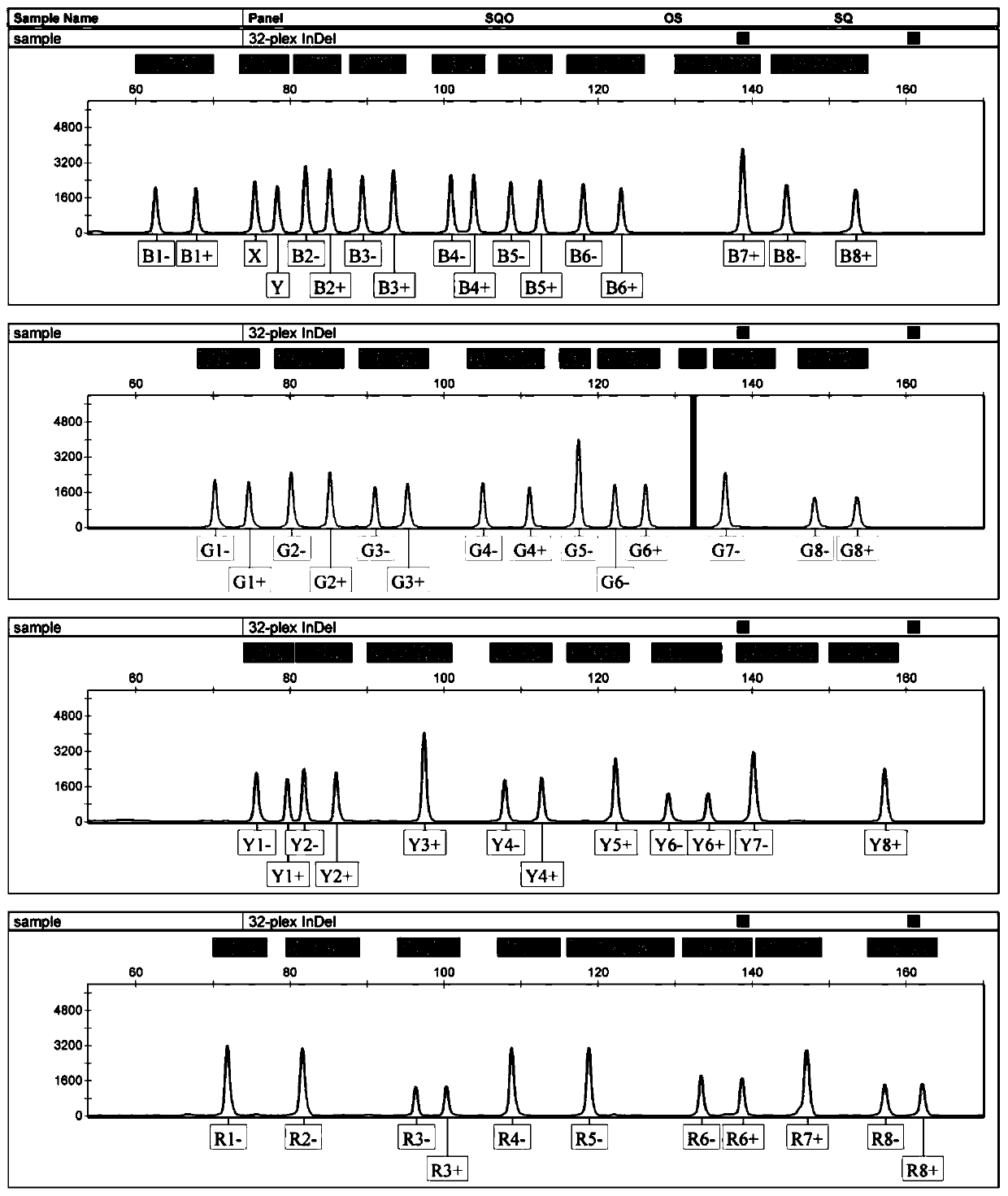
[0024] Example 1 Construction of 32-plex InDels multiplex amplification system
[0025] 1. DNA sample
[0026] Oral swabs or blood samples were collected from 204 unrelated individuals of Han nationality in southern China, including 78 females and 126 males. Genomic DNA was extracted by Chelex-100 method. DNA products were quantified using a Nanodrop 2000 spectrophotometer (ThermoScientific, USA). This study was approved by the Medical Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology.
[0027] 2. Selection of InDel markers and primer design
[0028] InDel markers were selected from 1000 Genomes Project data (the 1000Genomes Project) or published articles. All candidate InDel markers meet the following criteria: (1) indel markers are located on autosomes; (2) in the Chinese Han population, the minimum allele frequency (MAF) of each marker must be ≥ 0.4; (3) The insertion / deletion length of each InDel marker is 3-6bp, and a few of th...
Embodiment 2
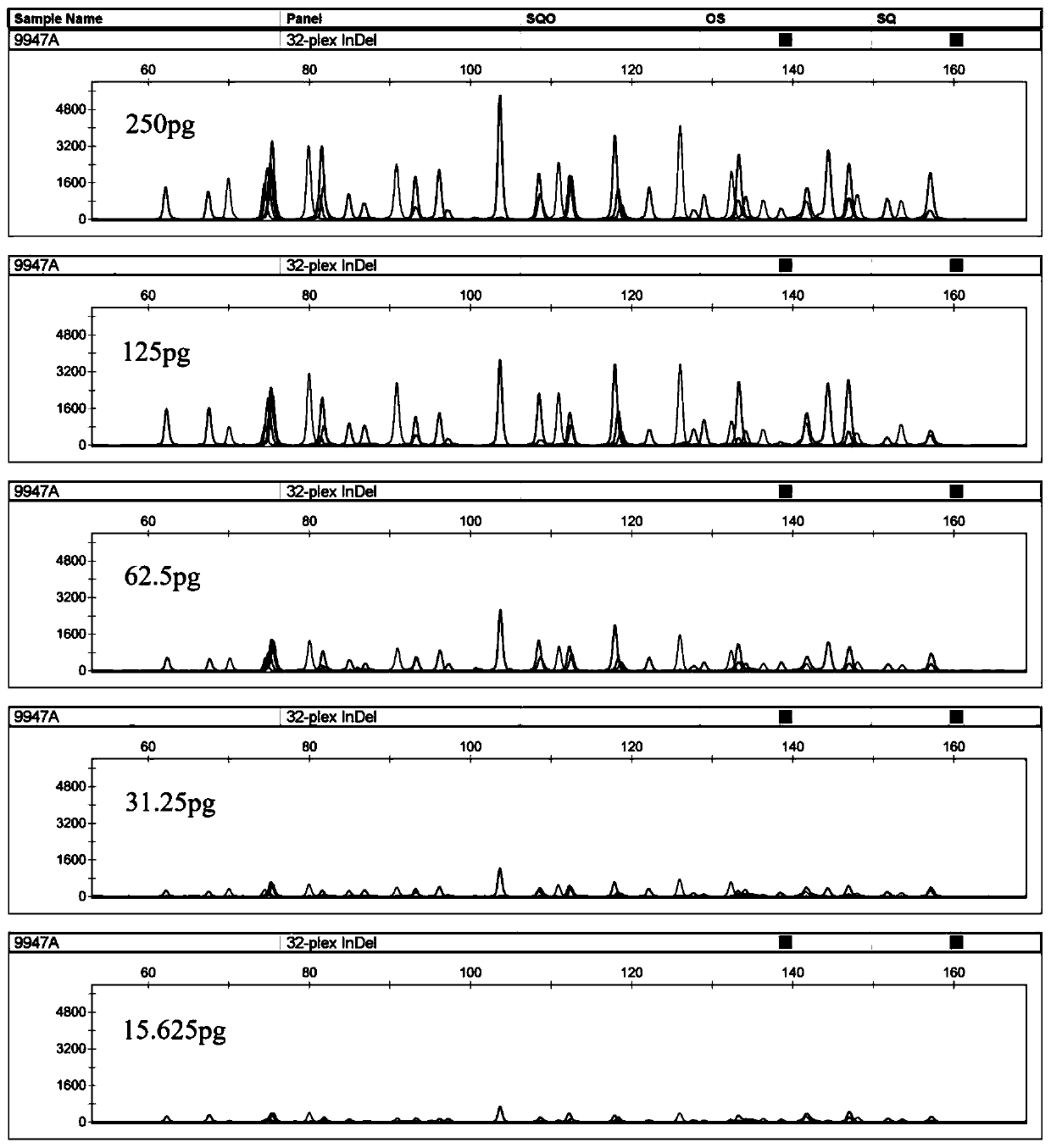
[0044] Example 2 Sensitivity Research and Degradation DNA Research
[0045] 1. Sensitivity
[0046] To determine the sensitivity of the 32-plex InDels Multiplex Amplification System, experiments were performed using control DNA 9947A. The control standard DNA 9947A was serially diluted to 500pg, 250pg, 125pg, 62.5pg, 31.25pg and 15.625pg, and PCR and capillary electrophoresis were performed according to the conditions in Example 1.
[0047] Typing map results such as figure 2 Shown (wherein the 500pg concentration DNA typing map is not shown). When the DNA amount is greater than or equal to 31.25pg, the complete typing result can be detected; while the DNA amount is 15.625pg, the peak height of a few sites is lower than the detection threshold. Although 31.25 pg of DNA is sufficient for detection, we recommend using DNA template amounts between 0.5-1 ng for PCR in forensic practice.
[0048] 2. Degrade DNA
[0049] In order to verify the ability of this method to detect ...
Embodiment 3
[0051] Example 3 Species Specificity Test
[0052] Multiplex amplification systems used in forensic applications also need to be specific for Homo genus. We used the Chelex-100 method to extract the DNA of five common animals at crime scenes, including rats, rabbits, pigs, cows, and chickens, for amplification and electrophoresis, and verified the 32-plexInDels multiplex amplification system.
[0053] The results showed that no amplified products were detected in rats, rabbits and chickens, while an insertion at 87 bp of a locus (rs3042783) was detected in DNA from cattle and pigs, see Figure 5 . After primer blast, it was confirmed that a pair of primers at rs3042783 could indeed amplify the DNA fragments of these two species. Overall, however, none of these animals were able to obtain complete DNA profiles using this multiplex amplification system, demonstrating that the 32-plex InDels multiplex amplification system can provide the species-specificity required for human D...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com