Method for removing and/or detecting nucleic acids having mismatched nucleotides
A nucleotide and nucleic acid technology, applied in biochemical equipment and methods, microbial assays/tests, ligases, etc., can solve the problems of not describing the main source of adaptor dimers, hybrid off-target cleavage, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach 1
[0078] Embodiment 1. A method of reducing adapter dimers comprising:
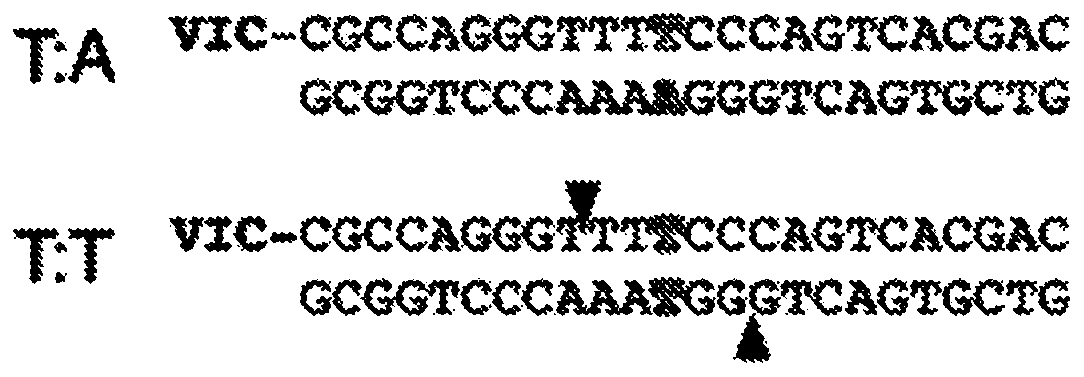
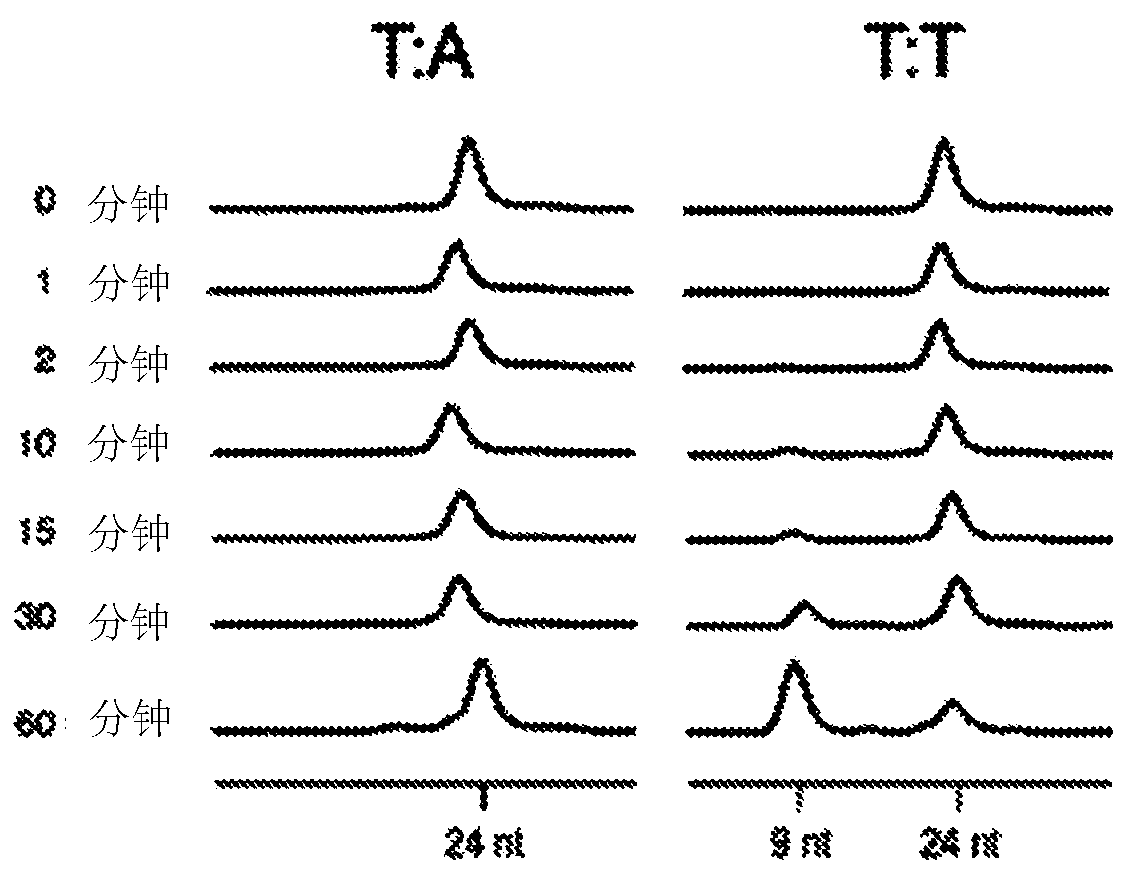
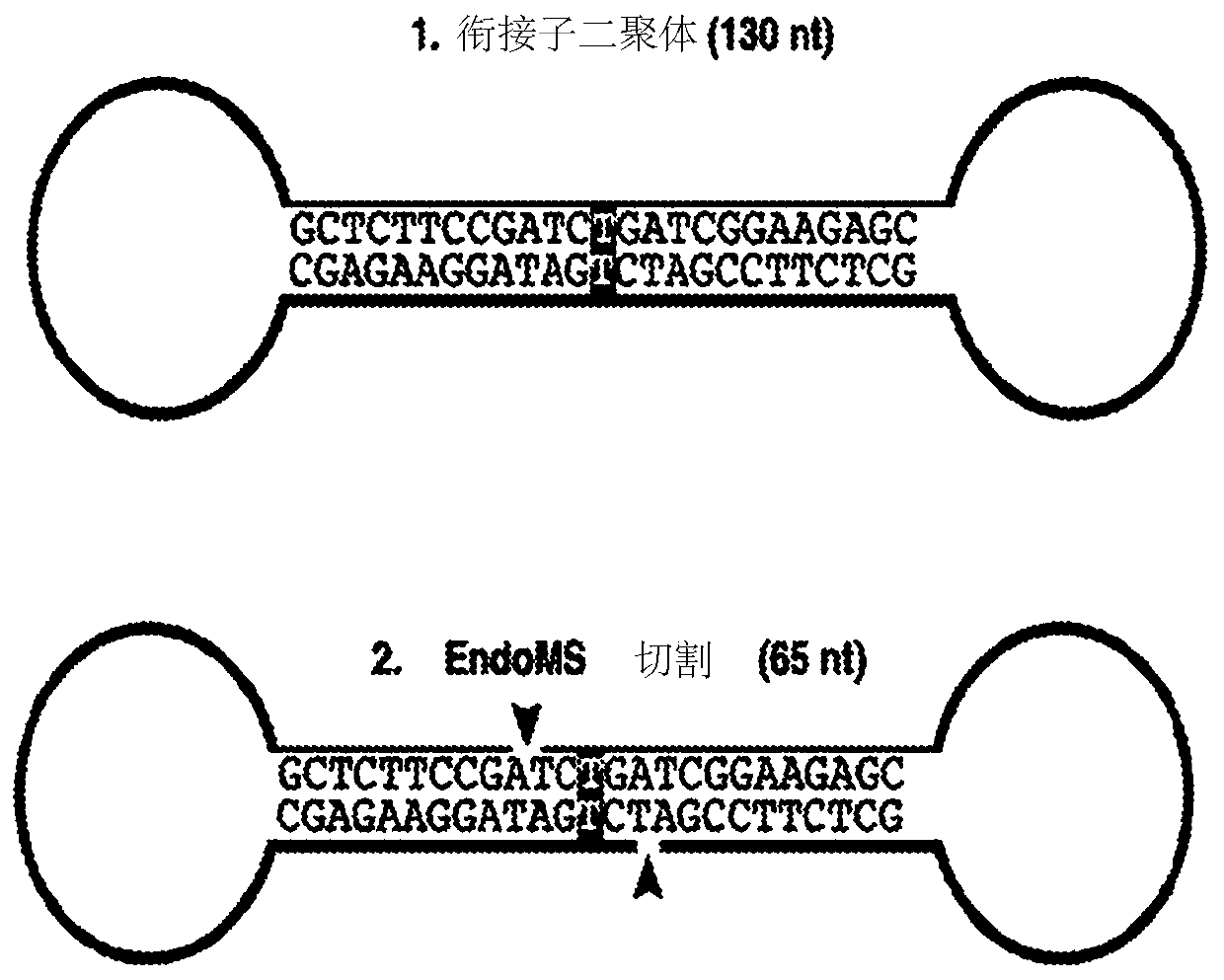
[0079] (a) Ligation of T-tailed double-stranded adapters to A-tailed double-stranded fragments of nucleic acids to generate dimerization of double-stranded adapters containing adapter-ligated fragments and T:T mismatches at ligation junctions body ligation products; and
[0080] (b) Cutting of both strands of the adapter dimer using EndoMS.
Embodiment approach 2
[0081] Embodiment 2. The method of embodiment 1, further comprising (c) amplifying the adapter-ligated fragments.
Embodiment approach 3
[0082] Embodiment 3. The method of embodiment 2, wherein amplification is performed using primers that hybridize to the adapter or its complement.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com