Identification method of Qingyu pig-derived components
A pig-derived, non-Qingyu technology, applied in the fields of molecular biology and food testing, can solve the problems of ineffective clustering of varieties, large sample size required for modeling, and large number of SNP sites, etc., to avoid counterfeit raw materials and products Harm to industry, enhancement of black pork value, effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0054] The preparation method of the template is as follows: mixing meat or meat products with sterilized deionized double-distilled water in a ratio of 1:3 to 1:5, and homogenizing it with a tissue homogenizer at 11000 to 13000 r / min for 6 to 10 minutes to make a uniform tissue. slurry. The total genomic DNA of the samples was extracted using a tissue DNA extraction kit.
[0055] Further, the 25 μL reaction system in the triple RT-PCR method is shown in Table 2:
[0056] Table 2
[0057]
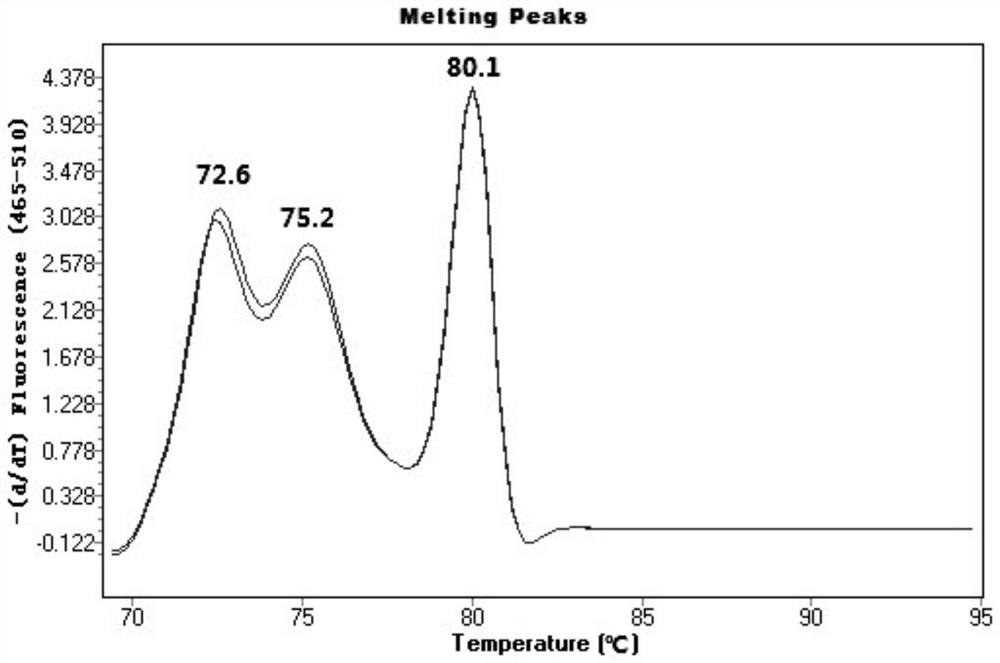
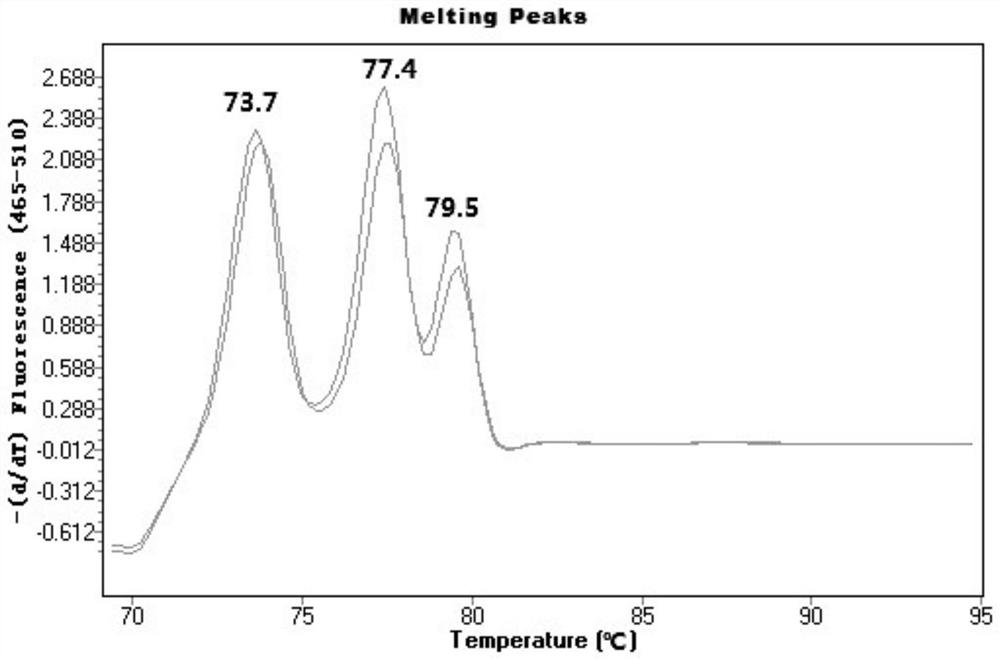
[0058] RT-PCR reaction conditions are: (1) Pre-denaturation: 95°C, 3min; (2) Denaturation: 95°C, 10-15s; Annealing and extension: 58-62°C, 20-45s, 30 cycles, (3) Melting curve preparation: 95°C, 1min, 70°C, 1min, heating to 95°C at a rate of 0.02°C / s, while continuously detecting the fluorescence intensity.
[0059] Considering the migration law of RT-PCR melting curve and the detection error of the instrument, a Tm value range is given for each primer to determine the result. Among ...
Embodiment 1
[0066] Example 1 The ability of SNP site combination to distinguish pig breeds
[0067] 1. Obtaining mitochondrial DNA sequence information
[0068] A total of 258 porcine mitochondrial genes were obtained by downloading from the NCBI website (https: / / www.ncbi.nlm.nih.gov / ) and sample sequencing, which were used to construct the identification method for Qingyu pigs. There were 172 pig mitochondrial genes, and 88 mitochondrial genes were obtained by DNA sequencing of pork samples, involving a total of 52 domestic and foreign pig breeds. For purchased pork samples, DNA was extracted using the DNeasy Animal Tissue DNA extraction kit. The extracted DNA was measured by fluorescence microplate reader at 260nm and 280nm, and the DNA concentration and purity were calculated. Then, mitochondrial gene information was obtained by PCR amplification and product sequencing.
[0069] 2. SNP site screening and theoretical accuracy
[0070] The collected 258 mitochondrial gene information wa...
Embodiment 2 3
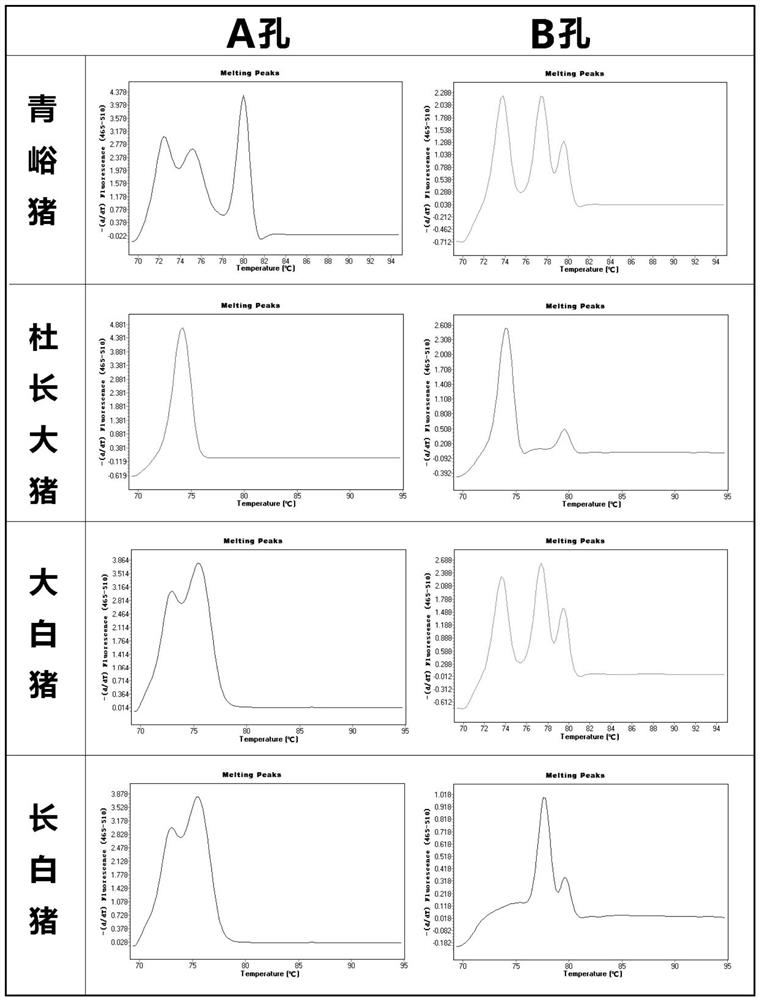
[0073] Example 2 Establishment and specificity of triple RT-PCR reaction system
[0074] 1. Sample processing method
[0075] Weigh 25mg Qingyu pig and 14 other common breed pork samples into centrifuge tubes respectively.
[0076] 2. DNA extraction method
[0077] Use the DNeasy animal tissue DNA extraction kit to extract DNA from each species, and operate according to the kit instructions, or other recognized extraction methods with the same efficacy. The extracted DNA was measured by fluorescence microplate reader at 260nm and 280nm, and the DNA concentration and purity were calculated.
[0078] 3. Primer sequence
[0079] A total of 9 SNP sites from SNP site 1 to SNP site 9 on the mitochondrial DNA of Qingyu pigs were used as the 3'-end targets of primers. A total of 36 pairs of primers were designed, and 6 pairs of specific primers were screened out: QY1-1F / R, QY1-2F / R, QY1-3F / R, QY2-1F / R, QY2-2F / R, QY2-3F / R. To improve the resolution and accuracy of the method, two ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com