Cloning, identification and application of salt tolerance related gene spliceosome
A technology for salt-tolerant genes and transgenic plants, which is applied in the cloning, identification and application of salt-tolerance-related gene splicing bodies, and can solve the problems of increasing the complexity of transcripts and proteomes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
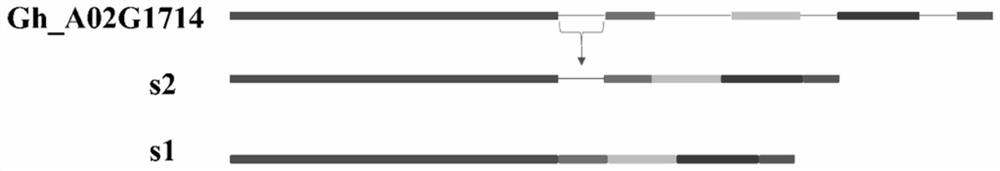
[0085] Embodiment 1, cloning and identification of spliced body
[0086] 1. Cloning of spliced bodies
[0087] 1. Extract the total RNA of 35 leaf tissues in the process, use the total RNA as a template, and use action primers F1: 5'-ATCCTCCGTCTTGACCTTG-3' and R1: 5'-TGTCCGTCAGGCAACTCAT-3' for PCR amplification to detect whether there is any genomic DNA residue . If PCR amplification yields a band of about 215 bp in size, there is genomic DNA remaining in the total RNA; otherwise, there is no genomic DNA remaining.
[0088] The result is as figure 1 As shown, CK is a PCR product obtained by using cDNA as a template and using action primer PCR amplification, and 1-8 is a PCR product obtained by using total RNA as a template and using action primer PCR amplification. The results showed that there was no genomic DNA residue in the total RNA extracted from leaves of Zhong35.
[0089] 2. Using reverse transcriptase to reverse transcribe the first-strand cDNA from Zhong 35 ...
Embodiment 2
[0108] Embodiment 2, embodiment 1 obtain the salt tolerance analysis of two shear bodies
[0109] 1. Construction of recombinant vector pBI121-s1
[0110] 1. Use the recombinant vector T-s1 as a template, and use F4: 5'-CTAGAGGATCCCCGGGATGGTTTCAGAAATTGGGGCTG-3' and R4: 5'-GATCGGGGAAATTCGAGCTCTTAAAAGTATTTCTGGAAGCATTCCTCTTC-3' as primers to perform PCR amplification to obtain a PCR fragment s1' with a size of about 2000 bp.
[0111] 2. Digest the vector pBI121 with restriction endonucleases XbaI and SacI, and recover the vector backbone 1 with a size of about 12.9 kb.
[0112] 3. Use the In-fusion HD Cloning Kit to connect the PCR fragment s1' and the vector backbone 1 to obtain the recombinant vector pBI121-s1.
[0113] The recombinant vector pBI121-s1 was double digested with XbaI and SacI, and the results were as follows: Figure 5 As shown in A, the recombinant vector pBI121-s1 produced two bands (lane 2) after double enzyme digestion, one of which was similar in size to u...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap