Pyrus betulaefolia transcription factor PbrWRKY40 and application of pyrus betulaefolia transcription factor PbrWRKY40 in increase of total acid content of plantand genetic improvement of salt resistance
A transcription factor, plant technology, applied in the field of genetic engineering, can solve problems such as limited
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Cloning of PbrWRKY40 Gene and Construction of Overexpression Vector
[0077] 1. RNA extraction
[0078] The research material, Du pear, was planted in the Pear Engineering Center of Nanjing Agricultural University, and its seedling age was 60 days. Select vigorously growing Du pear seedlings, randomly weigh 0.1g samples, and quickly freeze them with liquid nitrogen. RNA was extracted using the Total RNA Extraction Kit from Solebo, the specific method is as follows:
[0079] (1) Sample processing: Take 0.1g of fresh or -80°C frozen tissue, grind it in liquid nitrogen, add the powder to 1ml of lysate and mix well to obtain a homogenized sample;
[0080] (2) Place the treated sample at room temperature for 5 minutes, so that the nucleic acid protein complex is completely separated;
[0081] (3) Add 0.2 ml of chloroform to the homogenized sample after standing at room temperature, cover the tube cap, shake vigorously for 15 seconds, and place at room temperature for 3 to...
Embodiment 2
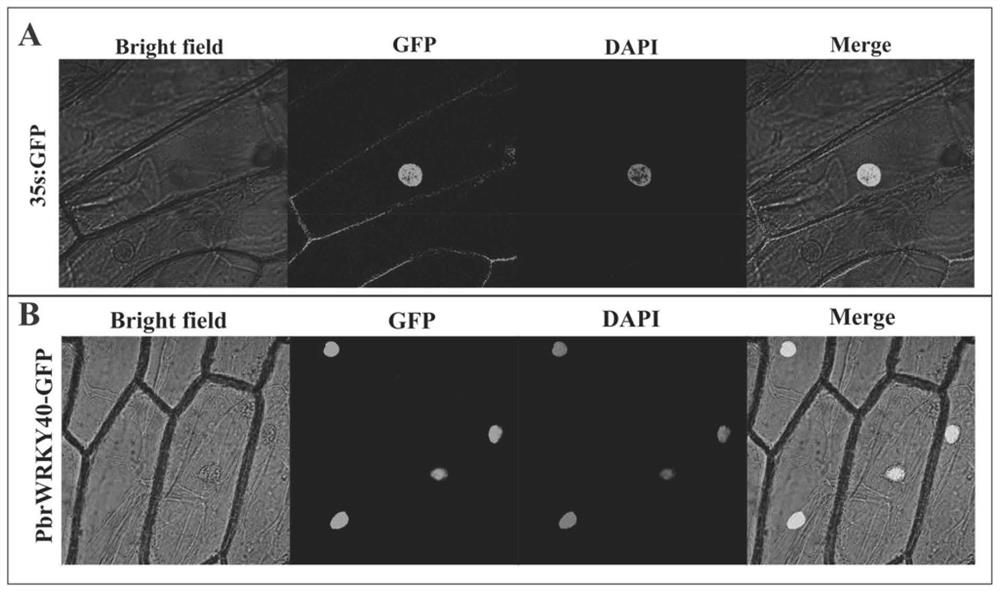
[0103] Subcellular localization of the gene encoding the transcription factor PbrWRKY40
[0104] According to the nucleotide sequence of the gene encoding the transcription factor PbrWRKY40 and the pJIT166-GFP vector map, Xba I and BamHI restriction sites were added before and after the gene sequence. The target gene extraction plasmid with correct sequencing results was used as a template, and amplified with primers (SEQ ID NO: 9, ATGGACTACTCAGCTGCATATGATG and SEQ ID NO: 10, ATAAGTATTATGTTGAAGTATTCTTCCTGATATG) with added restriction sites. The PCR program used was: 94°C pre-denaturation for 3 minutes ; Denaturation at 94°C for 30s, annealing at 58°C for 60s, extension at 72°C for 90s, 35 cycles; extension at 72°C for 10min. The stop codon TAG was removed from the 3' of the gene in order to allow the gene to be fused to GFP. After the PCR product was subjected to 1% agarose gel electrophoresis, the target band was recovered using a gel kit. The pJIT166-GFP vector plasmid was...
Embodiment 3
[0108] Genetic Transformation of Arabidopsis
[0109] 1. Plant transformation vector construction
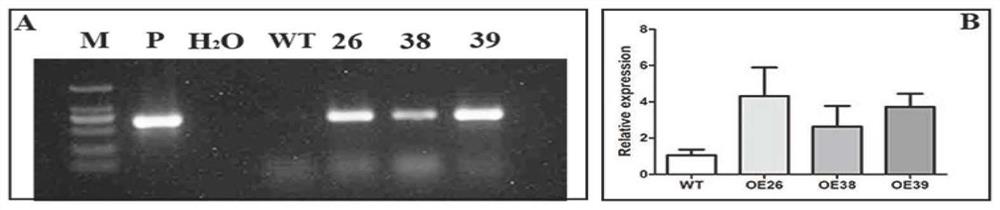
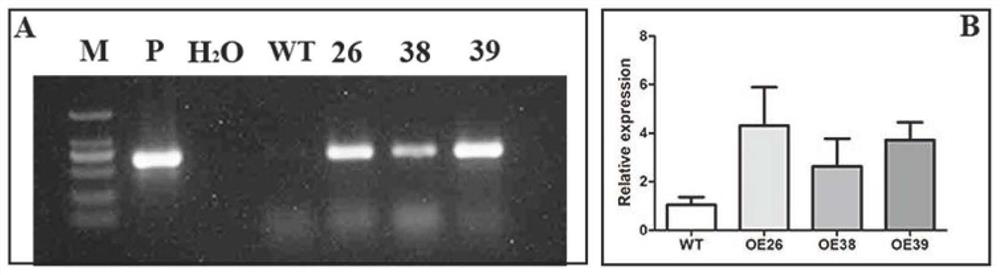
[0110] According to the multiple cloning site of the PCMBIA1300 vector and the coding region sequence of the PbrWRKY40 gene, the restriction site Xba I and BamH I were added, and the upper and lower PCR primers (SEQ ID NO: 9) were designed with primer primer 5.0 software according to the general principle of primer design and SEQ ID NO: 10). Using the clone of the PbrWRKY40 gene as a template, the PCR amplification is carried out with the PCR primers. The annealing temperature for PCR amplification was 58°C, and the PCR reaction system and amplification procedure were the same as those for PbrWRKY40 gene cloning. Gel purification was performed after amplification. The volume of the double enzyme digestion reaction of PCMBIA1300 vector is 40 μl, including: 10 μl of vector plasmid containing PCMBIA1300, 4 μl of 10×M buffer, 1 μl of Xba I and BamHI, and 24 μl of double distilled...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com