Engineered ph-dependent anti-CD3 antibodies and methods of production and use thereof
A technology of antibodies and antigens, applied in chemical instruments and methods, antibodies, anti-receptors/cell surface antigens/cell surface determinant immunoglobulins, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
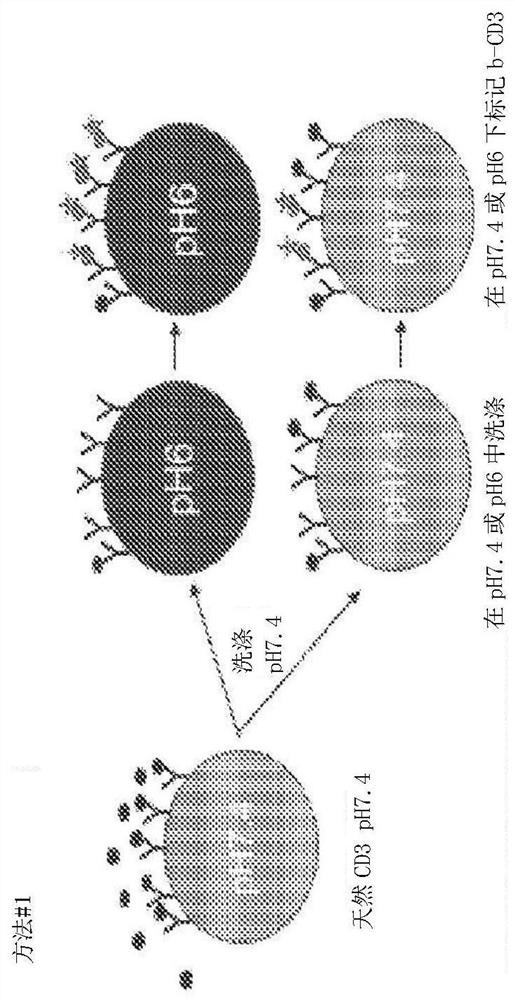
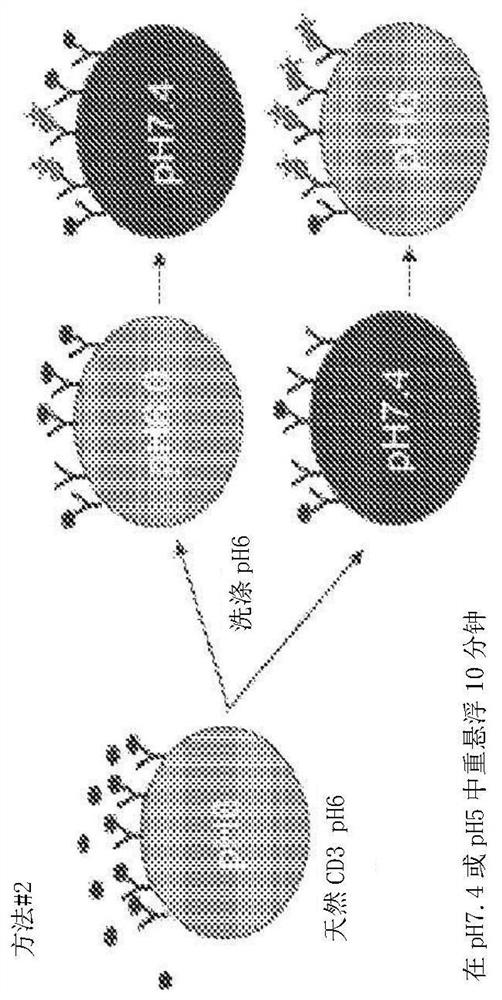
[0183] Example 1: Construction of an engineered pH-dependent CD3 library
[0184] The combinatorial histidine substitution library was derived from the parental anti-CD3 antibody clone ADI-26906 (antibody number 1 of Table 1). ADI-26906 was originally disclosed in PCT / US2018 / 031705, which is hereby incorporated herein by reference in its entirety (ADI-26906 is not pH engineered). Histidine incorporation was utilized using three library designs: 1) H3+L3 hopping double plus L1 single or double histidine (His) substitution (with and without NNK variegation adjacent to His), resulting in a theoretical diversity of 3.4x10 5 ;2) Premade H1 / H2 diversity library plus H3 skipping doublets, resulting in a theoretical diversity of 6.8x10 8 , and 3) H3+L3 NNK / His or His / NNK walking monomorphisms, resulting in a theoretical diversity of 1.2x10 5 . Libraries were generated and propagated as previously described (see eg, WO2009036379; WO2010105256; WO2012009568; Xu et al., Protein Eng Des...
Embodiment 2
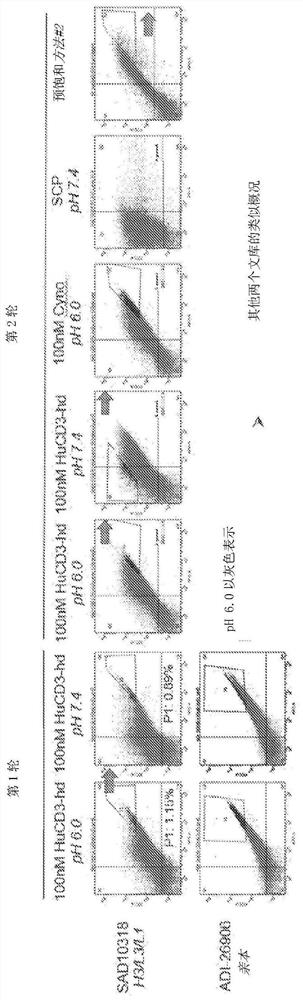
[0193] Example 2: Determining the affinity of anti-CD3 antibodies to CD3
[0194] By measuring its kinetic constant (k a 、k d 、K D ) to determine the affinity of anti-CD3 antibodies for CD3 at pH 6.0 and 7.4. ForteBio affinity measurements were generally performed as previously described (Estep et al., MAbs. 2013 5(2):270-8). Briefly, ForteBio affinity measurements were performed by online loading of antibodies (IgG) onto AHC sensors. The sensor was equilibrated offline for 30 minutes in assay buffer and then monitored online for 60 seconds for baseline establishment. For affinity binding measurements, sensors with loaded IgG were exposed to 100 nM antigen (human or cynomolgus CD3) for 3 minutes, after which they were transferred to assay buffer for 3 minutes for off-speed measurements. Kinetic data were fitted using a 1:1 binding model in the data analysis software provided by ForteBio. Table 2 provides kinetic constants for selected clones. Table 3 provides the equili...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com