Electromagnetic method and device for controlling single-chain nucleic acid perforating speed
A single-stranded nucleic acid, perforation speed technology, applied in the field of bioengineering, can solve problems that cannot be used in actual sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0015] Example 1: Control of perforation speed of BAC single-stranded DNA
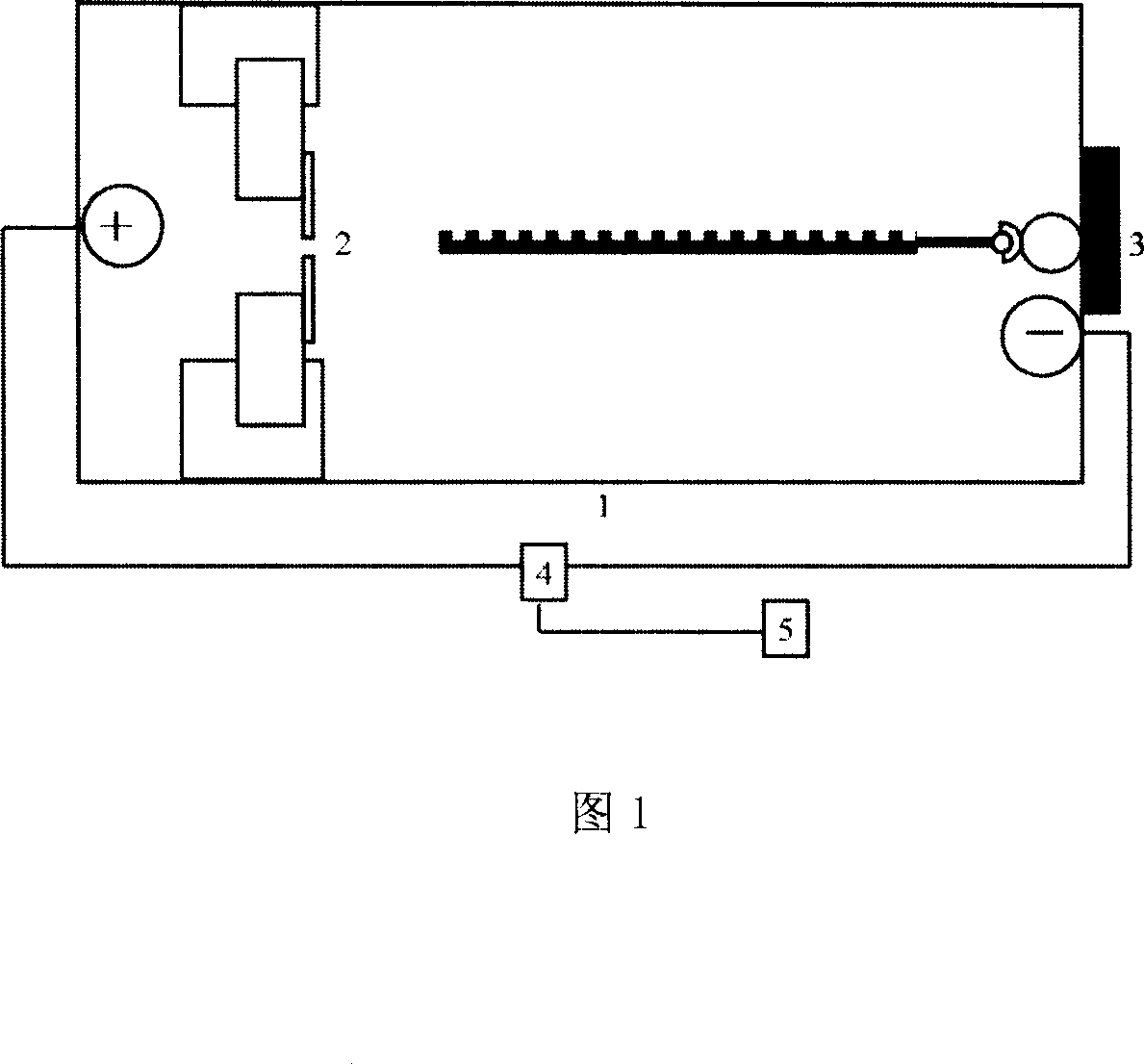
[0016] ①The complex connecting a single-stranded target DNA molecule and a magnetic bead is fixed on the negative pole of the electrophoresis tank with an electromagnet; the two poles of the electrophoresis tank are separated by nanopores.
[0017] ②Turn on the power supply, and give a pulling force (540pN) on the free end of the negatively charged target molecule to the positive pole. Since the end of the target molecule connected to the magnetic bead is fixed by the electromagnet (the electromagnetic force is 50pN), the result is that the target molecule is straightened. Then through fine-tuning, the electromagnetic force is weakened and the magnetic beads are released, but the electromagnet still attracts the magnetic beads, so that the target molecule moves slowly to the positive pole, and the speed of passing through the nanopore is controlled at 0.5 bases per millisecond, and the patch clamp To r...
Embodiment 2
[0019] Example 2: Control of the perforation speed of mRNA
[0020] ①The complex connecting a single-stranded target RNA molecule and a magnetic bead is fixed with an electromagnet on the negative pole of the electrophoresis tank; the two poles of the electrophoresis tank are separated by nanopores.
[0021] ②Turn on the power supply, and give a pulling force (1000pN) to the free end of the negatively charged target molecule towards the positive pole. Since the end of the target molecule connected to the magnetic bead is fixed by the electromagnet (the electromagnetic force is 1000pN), the result is that the target molecule is straightened. Then through fine-tuning, the electromagnetic force is weakened and the magnetic beads are released, but the electromagnet still attracts the magnetic beads, so that the target molecule moves slowly to the positive pole, and the speed of passing through the nanopore is controlled at 1 base per millisecond, and the patch clamp To record the ...
Embodiment 3
[0023] Example 3: Control of perforation speed of cosmid single-stranded DNA
[0024] ① The complex connecting a cosmid single-stranded RNA molecule and a magnetic bead is fixed with an electromagnet at the negative pole of the electrophoresis tank; the two poles of the electrophoresis tank are separated by nanopores.
[0025] ②Turn on the power supply, and give a pulling force (500pN) to the free end of the negatively charged target molecule towards the positive pole. Since the end of the target molecule connected to the magnetic bead is fixed by the electromagnet (the electromagnetic force is 500pN), the result is that the target molecule is straightened. Then through fine-tuning, the electromagnetic force is weakened and the magnetic beads are released, but the electromagnet still attracts the magnetic beads, so that the target molecule moves slowly to the positive pole, and the speed of passing through the nanopore is controlled at 1 base per millisecond, and the patch clam...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com