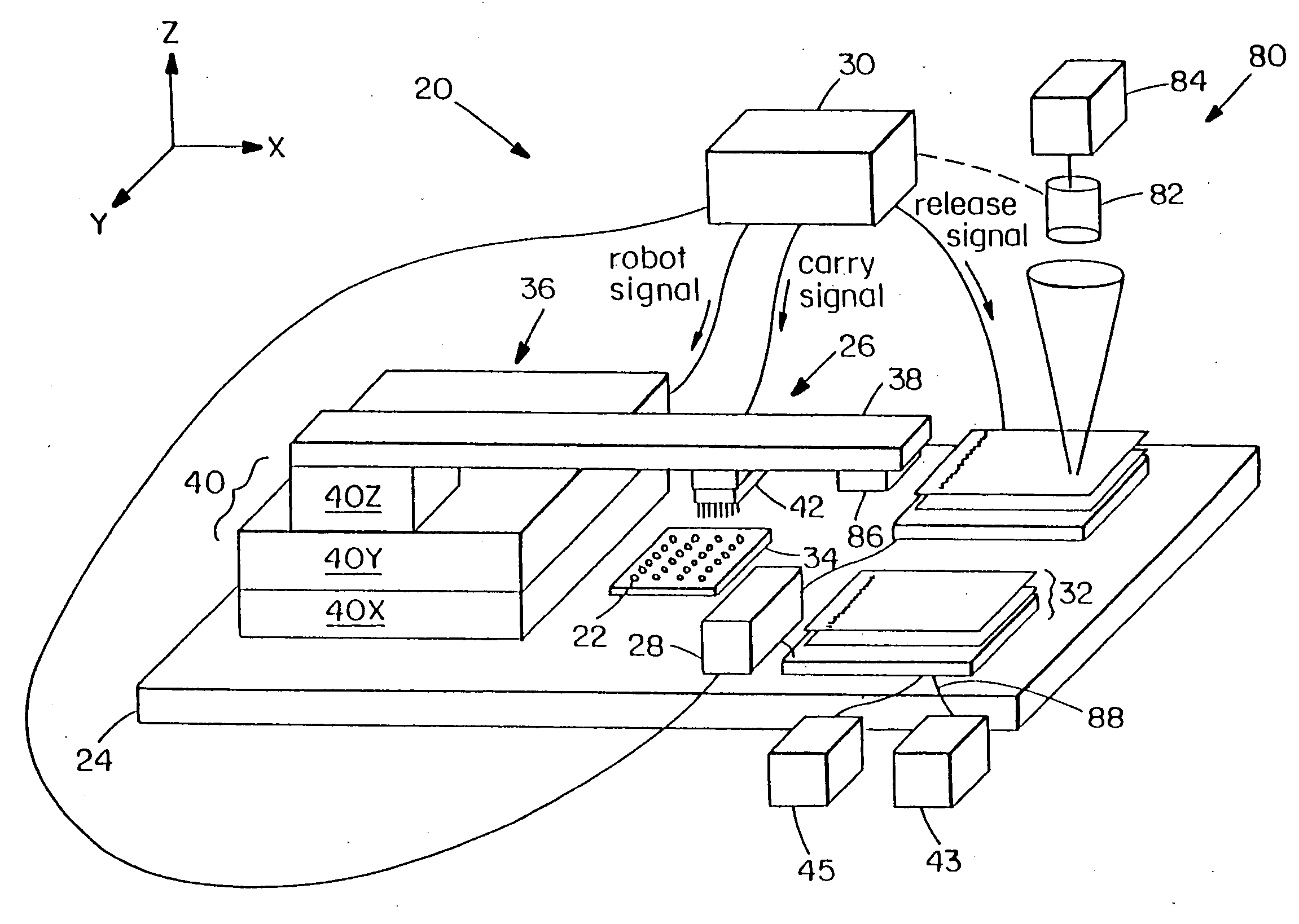
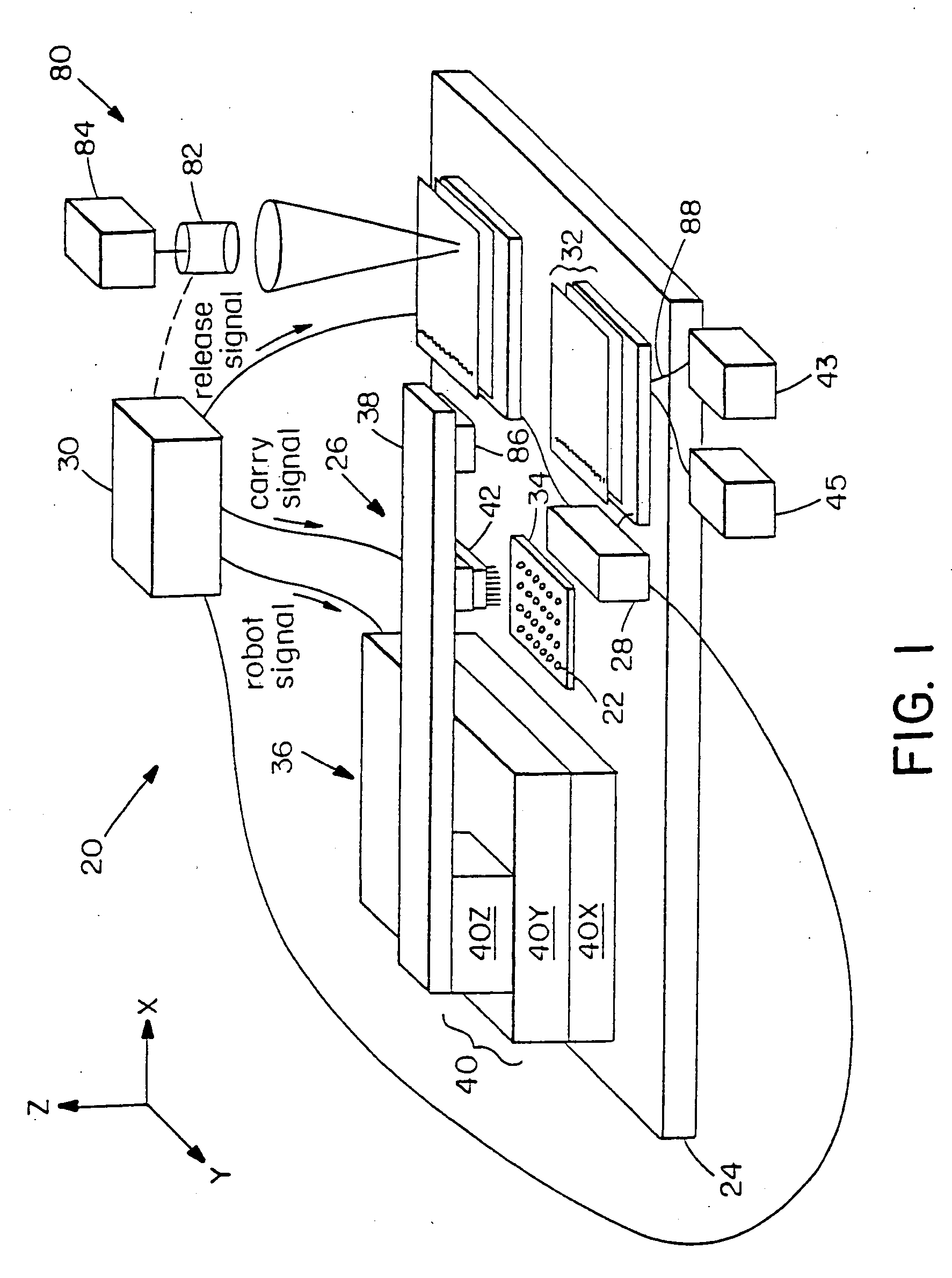
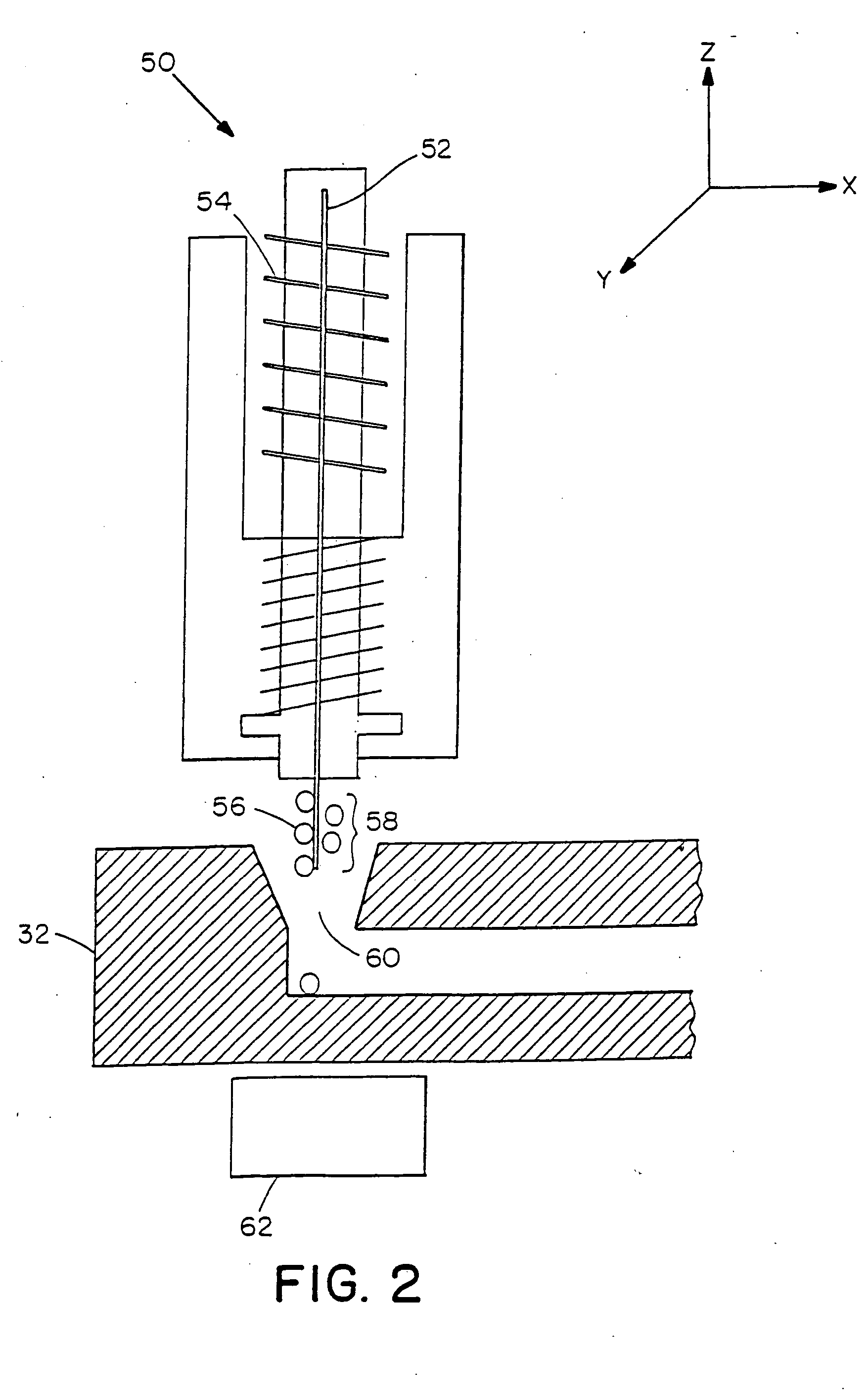
Methods and apparatus for processing a sample of biomolecular analyte using a microfabricated device
a microfabricated device and biomolecular analyte technology, applied in the direction of fluid pressure measurement, liquid/fluent solid measurement, peptide, etc., can solve the problems of increasing the speed of str analysis, significant decreases in electrophoretic run-time, etc., to achieve baseline-resolved electrophoretic separation of single-locus str samples, improve speed, and improve the effect of speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Determination of Factors Which Influence the Speed of STR Analysis
[0130] Initially, the general operation of the microfabricated device for genotyping by STR analysis was characterized. The objective was to determine the factors which influence the ultimate speed of such analyzes in microchip devices. Device performance was followed for the STR system CTTv, which consists of the four loci CSF1PO, TPOX, THO1 and vWA, each of which contains STR alleles which differ in length by four base pairs. The four loci, CSF1PO, TPOX, THO1 and vWA, contain 9, 5, 7, and 8 common alleles respectively. FIG. 12A shows the separation of the CTTv ladder ranging from 140 to 330 bases by microchip gel electrophoresis using one of our devices. This ladder was used as an internal sizing standard for the allelic profiling. In FIG. 12A the alleles of all the four loci are well resolved in less than two minutes with measured resolution R,
R=([2 ln 2]1 / 2)(t2−t1) / (hw1+hw2), (1)
where tn is the retention tim...
example 2
Assay of PCR Amplified Samples
[0136] The microchip device was used for genotyping of PCR amplified samples of eight individuals which were spiked with the CTTv ladder as the internal size standard and assayed on the microchip gel system. In all eight cases, the alleles could be identified with no ambiguity in under two minutes and the results were in complete agreement with data produced by traditional slab gel electrophoresis, which typically required 80, 94, 112 and 143 min to detect and resolve the alleles of vWA, THO1, TPOX, and CSF1PO respectively. The spiking experiment for one of the individuals is shown in FIG. 12B. The individual is clearly homozygous for vWA (allele 14) and heterozygous for THO1 (alleles 7 / 9), TPOX (alleles 8 / 9) and CSF1PO (alleles 10 / 14).
[0137] Results, thus, have demonstrated that the quadruplex STR system CTTv can be analyzed with high accuracy in less than two minutes and a single locus in 30 seconds by microchip gel electrophoresis. Compared to capi...
example 3
Evaluation of the Feasibility of Loading Schemes
[0142] A glass microchip with laser-drilled injection ports leading to microchannels was interfaced with a 1 cm thick gasket fabricated from polydimethylsiloxane (PDMS) silicone elastomer material (Dow Coming Sylgard 184, Schenectady, N.Y.). Capillaries of inner diameter 100 μm and outer diameter 375 μm were molded into the gasket and set in place while the PDMS cured to a semi-rigid final state. No rigid support structure was used in these trials, and the capillary-gasket system was aligned to the injection holes on the microchip manually with the aid of a microscope. The free end of the 10 cm long capillary was inserted into a container of fluorescein and a voltage of 50 volts per centimeter was applied between the container and the microchip channels. The microchip was illuminated with an argon ion laser source operating at 488 nm, and the fluorescing fluorescein was observed flowing from its original container to the microchannels...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| angle | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com