Methods And Compositions For Transcription-Based Nucleic Acid Amplification
a transcription-based nucleic acid and amplification technology, applied in the field of polynucleotide amplification, can solve the problems of limiting affecting the sensitivity of the method, and affecting the applicability of the method,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
The First Transcription Module of Composite Primer-Based Methods
[0265]The general methods that are described in this example are also utilized in other examples of the first transcription module of composite primer-based methods provided herein.
Buffers
[0266]Buffers that were used throughout the composite primer-based examples are made with the following materials.
[0267]TE Buffer: 10 mM Tris-HCl, pH 8.0, 1 mM EDTA
[0268]TBE Buffer: 89 mM Tris base, 89 mM boric acid, 2 mM EDTA, pH 8.3
[0269]FX Buffer: 20 mM Tris-SO4, pH 9.0, 20 mM (NH4)2SO4, 0.1% NP-40
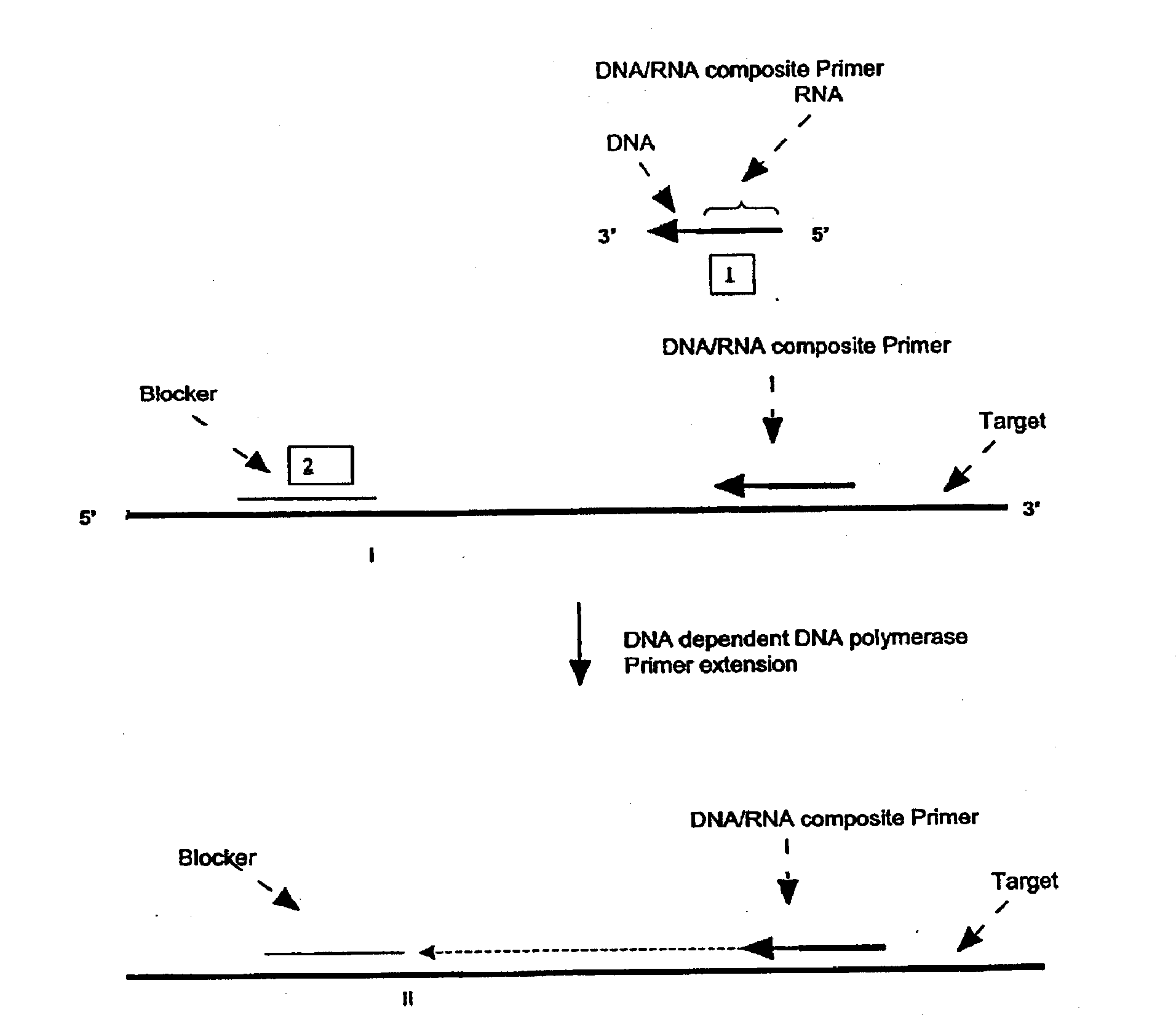
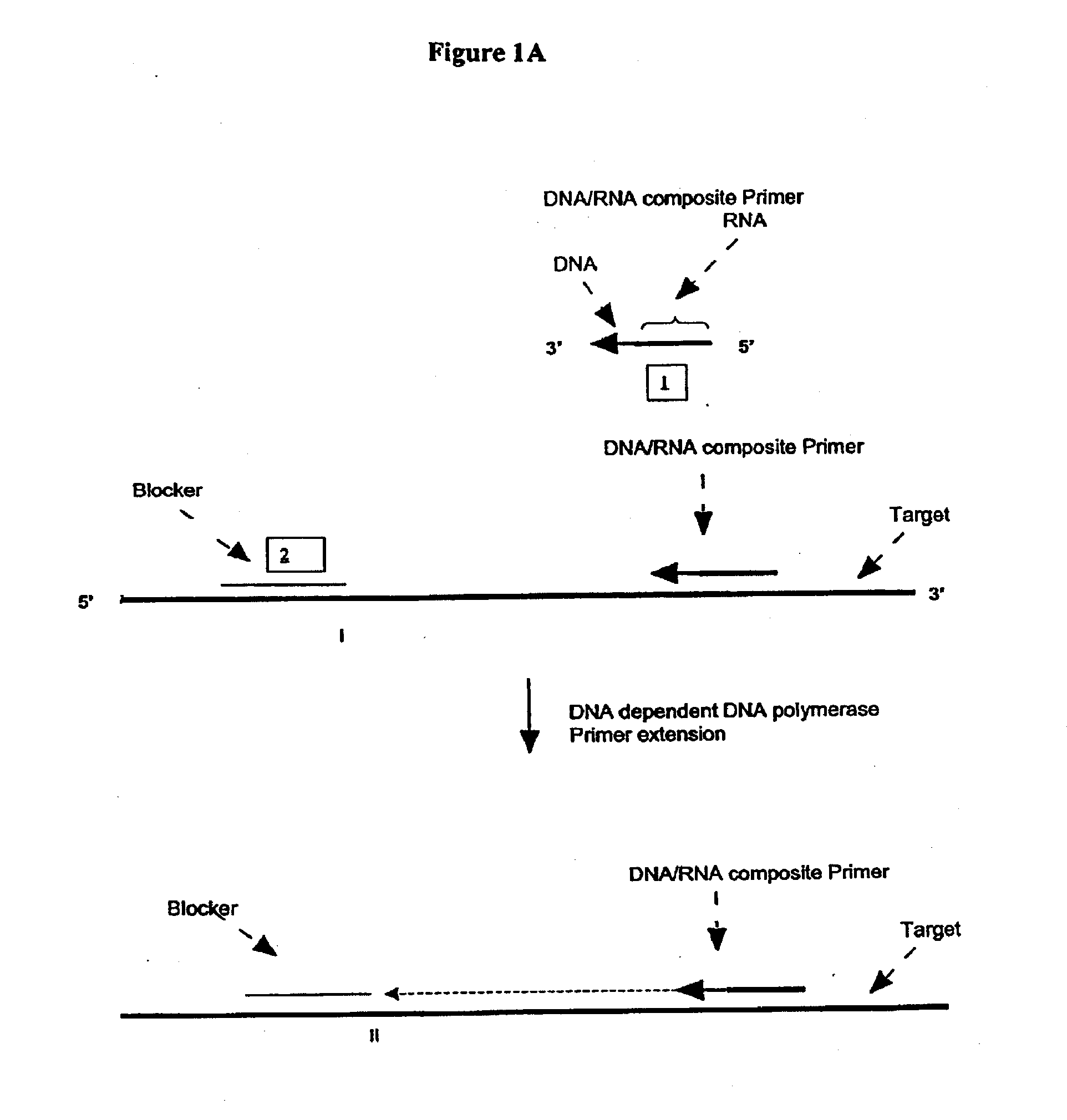
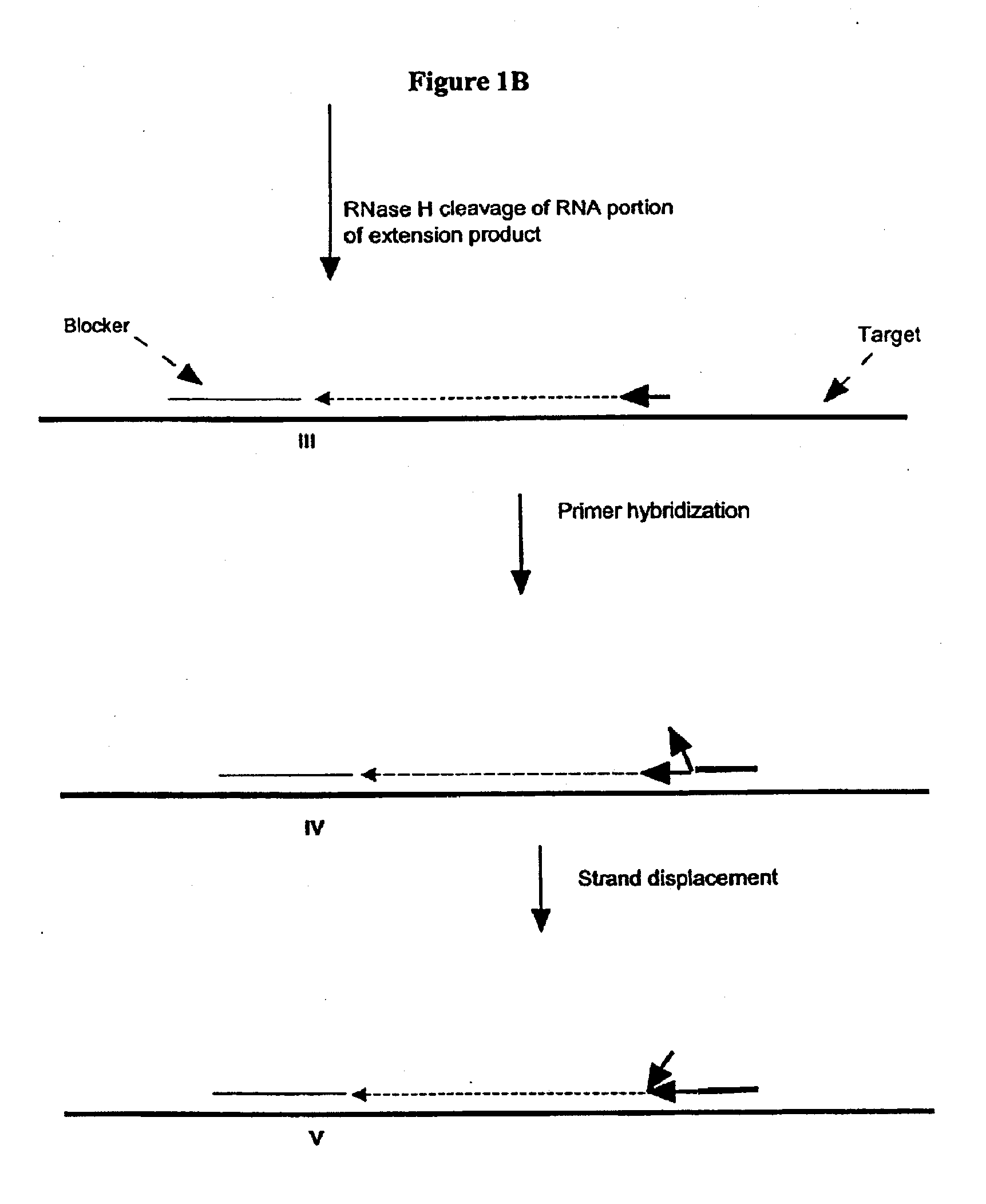
A First Illustration of the First Transcription Module of Amplification Methods Using a Single Composite Primer, DNA Polymerase and Rnase H
[0270]Sequence amplification was performed in 15 μl reactions containing 20 mM Tris-HCl, pH 8.5, 6.0 mM MgCl2, 1.0 mM dATP, 1.0 mM dCTP, 1.0 mM dTTP, 0.8 mM dGTP, 0.2 mM dITP (dNTP's from Amersham), 0-6% DMSO, 0-8% glycerol, 0-100 ug / ml acetylated BSA (Ambion, Austin, Tex.), 0.6 Units / μl recombinant rib...
example 2
A Second Illustration of the First Transcription Module of a Composite Primer-Based Amplification Method
[0281]Amplification using a single composite primer, DNA polymerase, RNase H, and TSO or blocker was performed. Reaction mixtures containing all reaction components, as described above, as well as reaction mixtures without one of the key reagents such as composite primer, RNase H, or Bca DNA polymerase (Panvera, Madison, Wis.) were spiked with 1010 copies of synthetic ssDNA target (IA010—sequence listed in Example 1). A negative control reaction containing all the reagents and no target ssDNA, was also included. The amplification of target DNA sequence was carried out as described above. Target denaturation was carried out at 70° C. and the isothermal amplification was carried out at 65° C.
[0282]The amplification products were resolved by gel electrophoresis. No amplification products were detected in reaction mixtures without primer, RNase H or Bca DNA polymerase.
[0283]Probe IA01...
example 3
Promoter Template Oligonucleotide-Based First Transcription Module of a Composite Primer-Based Amplification Method
[0284]The promoter-template oligonucleotide (PTO) contains two essential sequence motifs: a T7 promoter sequence (5′-TAATACGACTCACTATAGGGAgGAG)) and a sequence complementary to the ssDNA template. Four versions of a PTO were designed (IA012, IA012b, IA015, IA015b). IA012 PTO is a 67-mer and has a sequence of: GGAATTCTAATACGACTCACTATAGGGAGAGATCGAGTAGCTCCGGTACGCTGATCAAAGATCCGTG. IA012 PTO contains two sequences in addition to the core T7 promoter: a 5′-extension (5′-GGAATTC) and a spacer (5′-ATCGAGTAGCTC) between the promoter and the target DNA-complementary sequence. IA015 is the shorter PTO (48-mer), lacking both the 5′-extension and the spacer. IA015 PTO has the sequence of: TAATACGACTCACTATAGGGAGAGCGGTACGCTGATCAAAGATCCGTG. IA012b PTO is a 60-mer which contains the spacer, but not the extension. IA012b PTO has the sequence: TAATACGACTCACTATAGGGAGAGATCGAGTAGCTCCGGTACGCT...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com