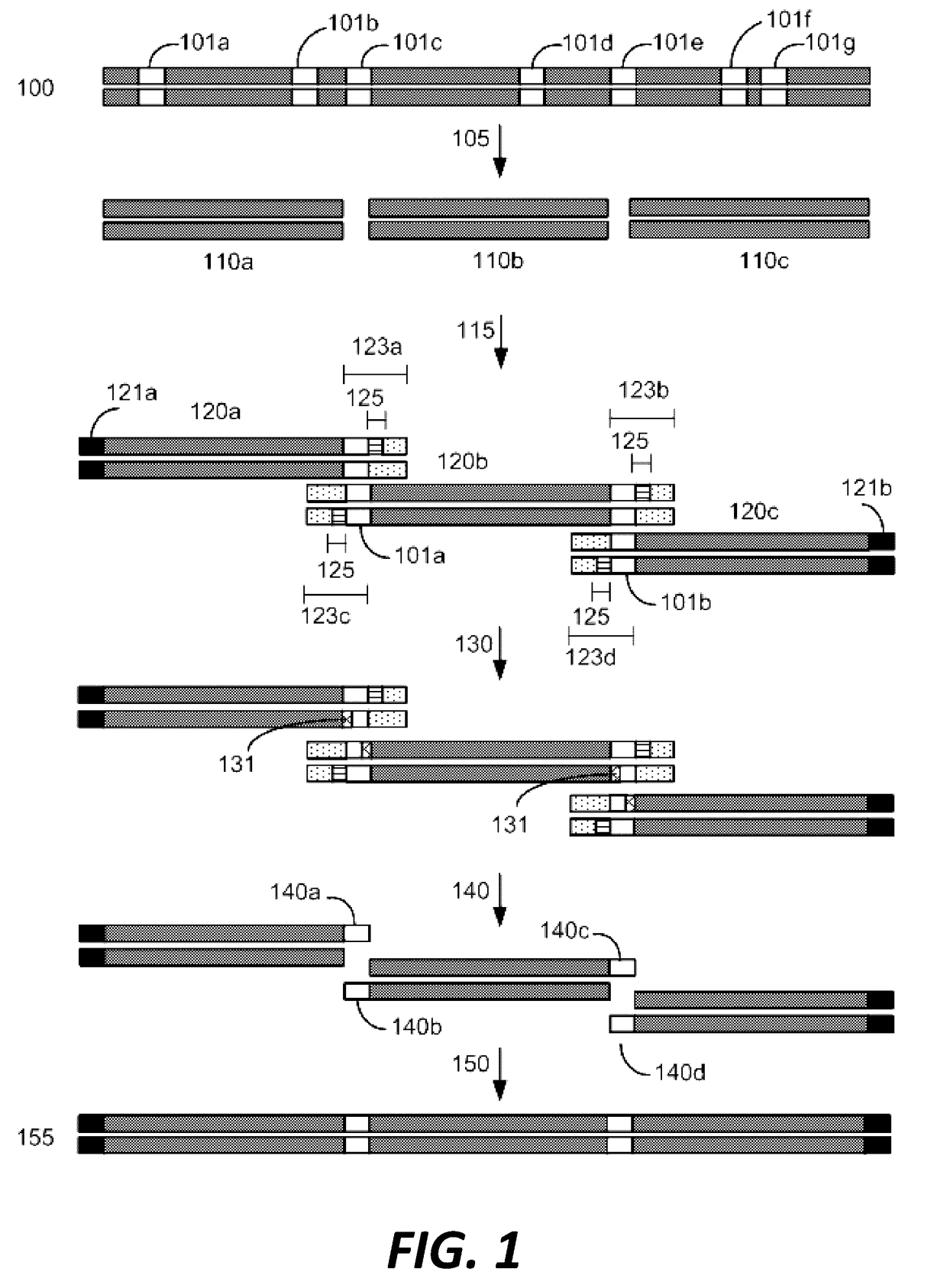
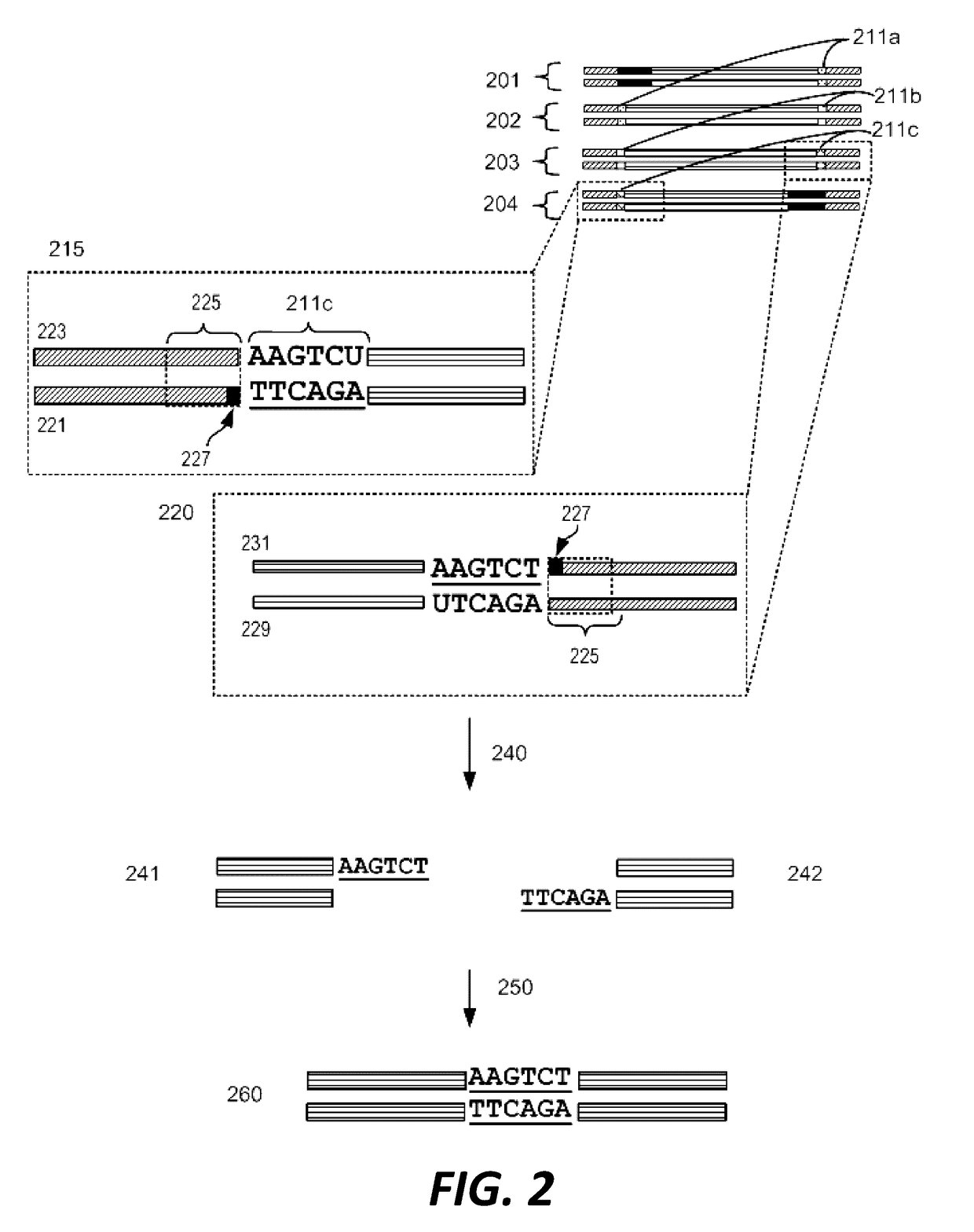
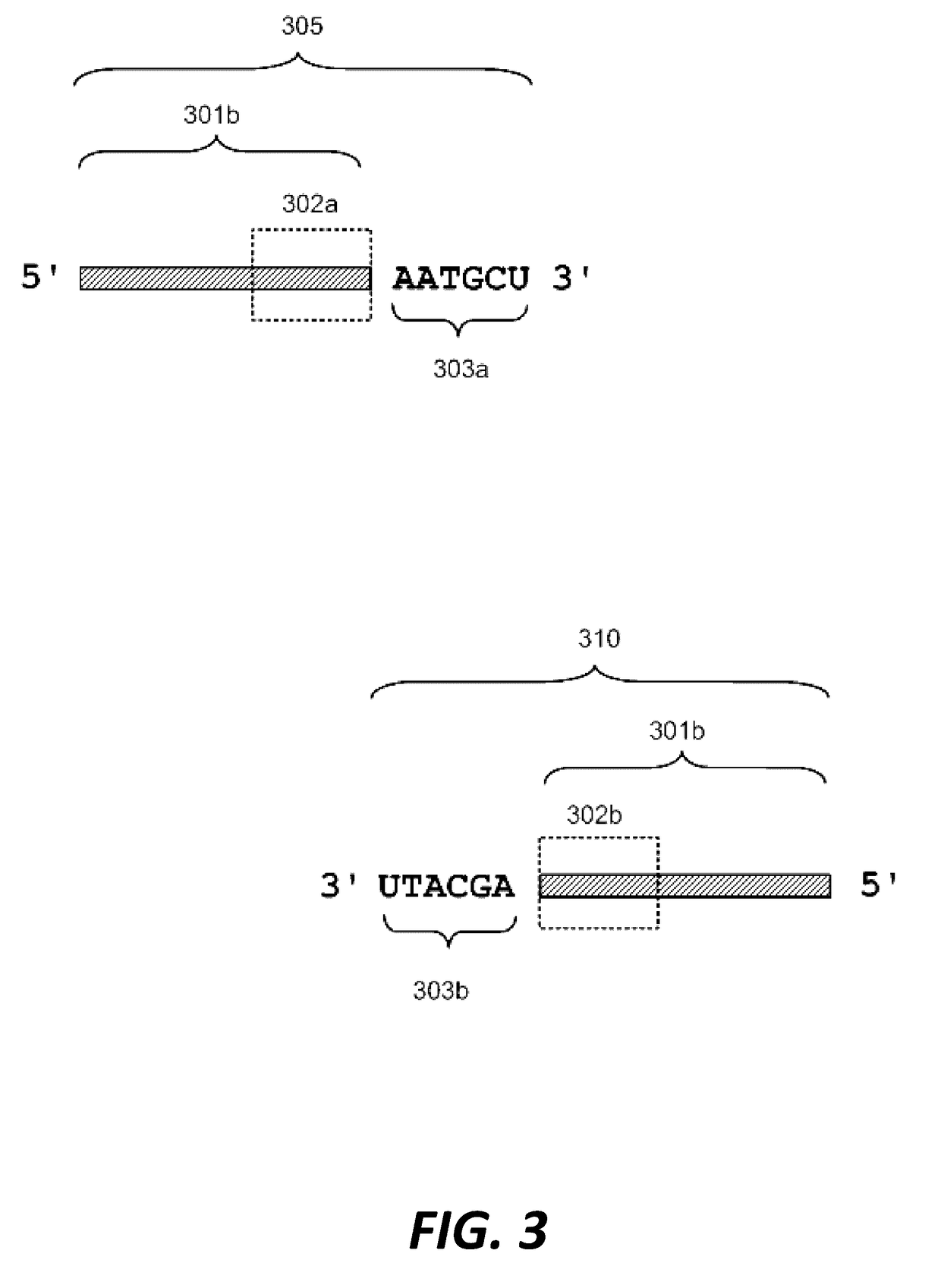
Compositions and methods for synthetic gene assembly
a technology of synthetic genes and assembly methods, applied in the field of synthetic gene assembly, can solve the problems of scalability, automation, speed, accuracy, cost, and limited assembly of nucleic acids from shorter segments, and achieve the effect of high fidelity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
on Enzyme-Free Ligation of a Gene Fragment Using Sticky Ends
[0150]Amplification with Uracil-Containing PCR Primers
[0151]A gene of about 1 kB (the “1 kB Gene Construct”) was selected to perform restriction enzyme-free ligation with a vector:
(SEQ ID NO.: 48)5′CAGCAGTTCCTCGCTCTTCTCACGACGAGTTCGACATCAACAAGCTGCGCTACCACAAGATCGTGCTGATGGCCGACGCCGATGTTGACGGCCAGCACATCGCAACGCTGCTGCTCACCCTGCTTTTCCGCTTCATGCCAGACCTCGTCGCCGAAGGCCACGTCTACTTGGCACAGCCACCTTTGTACAAACTGAAGTGGCAGCGCGGAGAGCCAGGATTCGCATACTCCGATGAGGAGCGCGATGAGCAGCTCAACGAAGGCCTTGCCGCTGGACGCAAGATCAACAAGGACGACGGCATCCAGCGCTACAAGGGTCTCGGCGAGATGAACGCCAGCGAGCTGTGGGAAACCACCATGGACCCAACTGTTCGTATTCTGCGCCGCGTGGACATCACCGATGCTCAGCGTGCTGATGAACTGTTCTCCATCTTGATGGGTGACGACGTTGTGGCTCGCCGCAGCTTCATCACCCGAAATGCCAAGGATGTTCGTTTCCTCGATATCTAAAGCGCCTTACTTAACCCGCCCCTGGAATTCTGGGGGCGGGTTTTGTGATTTTTAGGGTCAGCACTTTATAAATGCAGGCTTCTATGGCTTCAAGTTGGCCAATACGTGGGGTTGATTTTTTAAAACCAGACTGGCGTGCCCAAGAGCTGAACTTTCGCTAGTCATGGGCATTCCTGGCCGGTTTCTTGGCCTTCAAACCGGACAGGAATGCCCAAGTTAACGGAAAAACC...
example 2
of LacZ Gene into a Plasmid
[0157]A LacZ gene was assembled into a 5 kb plasmid from three precursor LacZ fragments and 1 precursor plasmid fragment. Assembly was performed using 9 different reaction conditions.
[0158]Preparation of Precursor Plasmid Fragments
[0159]A 5 kb plasmid was amplified with two different sets of primers for introducing a sticky end motif comprising a non-canonical base (SEQ ID NO.: 53): set A (SEQ ID NOs.: 54 and 55) and set B (SEQ ID NOs.: 56 and 57), shown in Table 6, to produce plasmid precursor fragments A and B, respectively.
TABLE 6Sequence identities of plasmid primers.SequencePrimeridentitynameSequenceSEQ IDplasmid-FaTGATCGGCAATGATATG / ideoxyU / CTGNO.: 54GAAAGAACATGTGSEQ IDplasmid-RaTGATCGGCAATGATGGC / ideoxyU / TATNO.: 55AATGCGACAAACAACAGSEQ IDplasmid-FbTGATCGGCAATGATATG / ideoxyU / CGCNO.: 56TGGAAAGAACATGSEQ IDplasmid-RaTGATCGGCAATGATGGC / ideoxyU / CGTNO.: 57ATAATGCGACAAACAAC
[0160]Each primer set comprises, in 5′ to 3′ order: 6 adaptor bases (TGATCG, SEQ ID NO.: 5...
example 3
torial Target Nucleic Acid Library
[0169]An enzyme of interest having an activity to be improved is selected. Specific amino acid residues relevant to enzyme activity and stability are identified. The nucleic acid sequence encoding the enzyme is obtained. Bases corresponding to the specific amino acid residues are identified, and the nucleic acid is partitioned into fragments such that each fragment spans a single base position corresponding to a specific amino acid residue.
[0170]Target nucleic acid fragments are synthesized such that identified bases corresponding to the specific amino acid residues are indeterminate. Target nucleic acid fragments are amplified using a uridine primer and treated with a sequence adjacent nick enzyme and a uridine-specific nick enzyme. Cleaved end sequence is removed and target nucleic acid fragments are assembled to generate a target nucleic acid library. Aliquots of the library are sequenced to confirm success of the assembly, and aliquoted molecule...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com