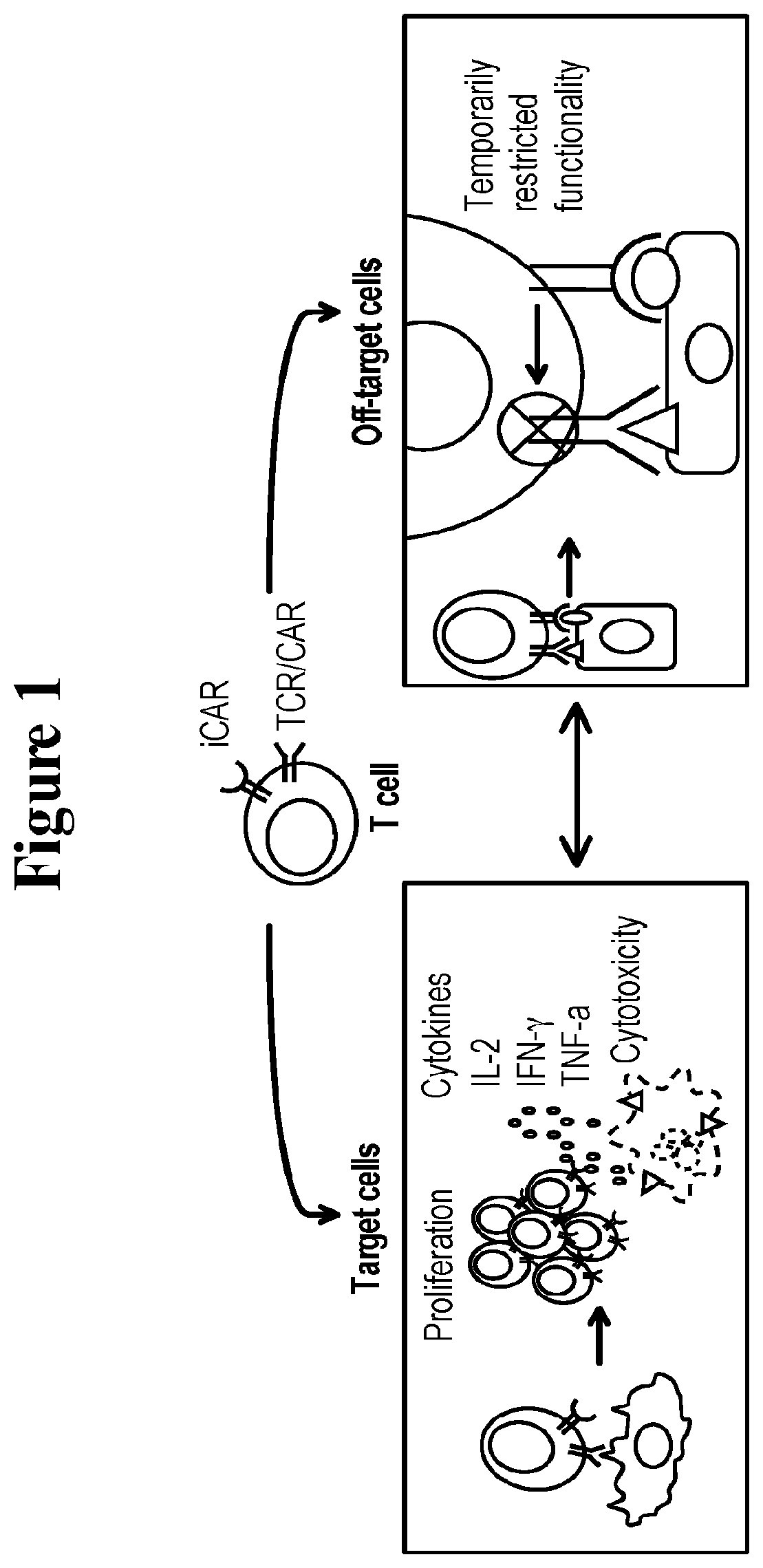
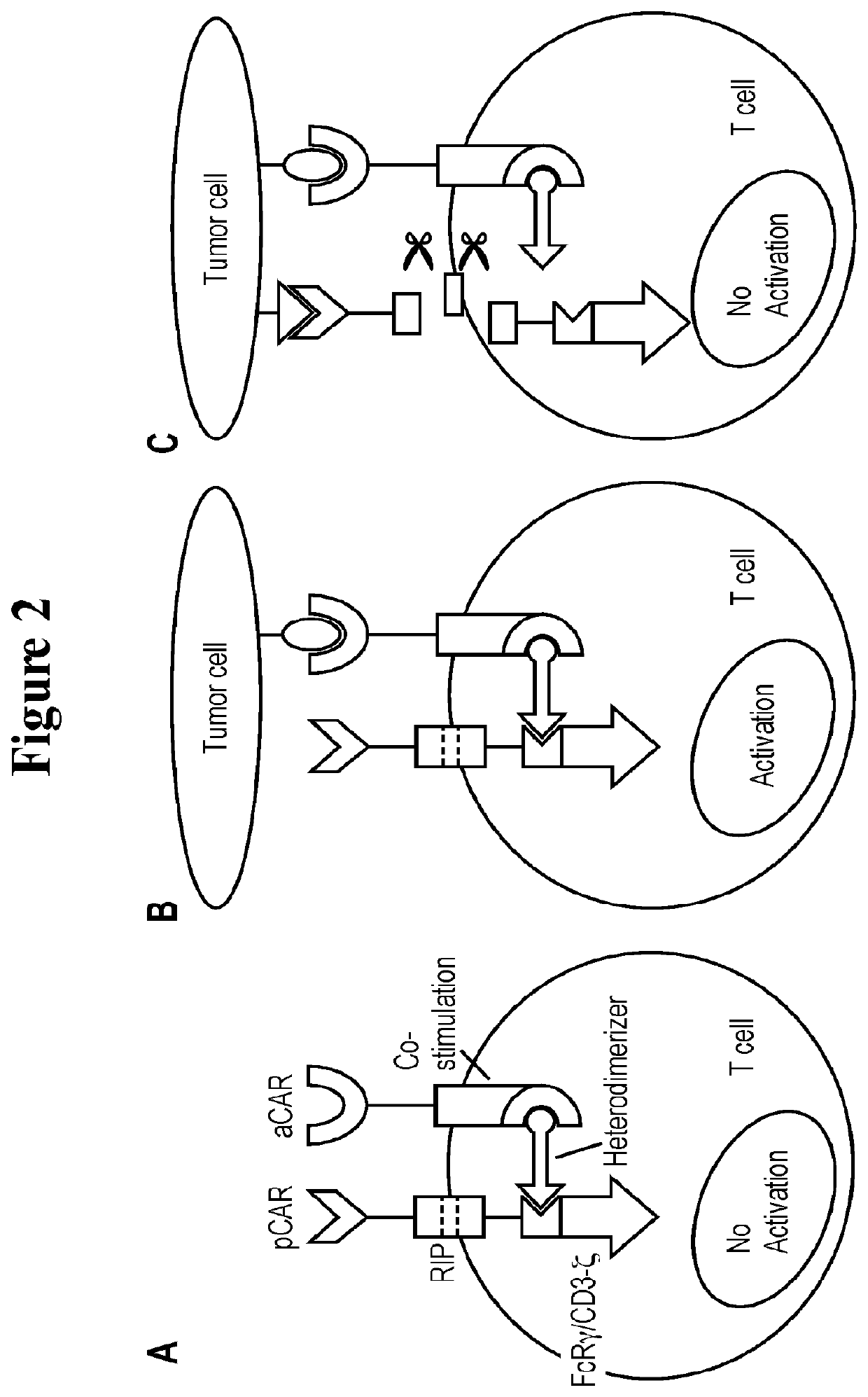
Universal platform for car therapy targeting a novel antigenic signature of cancer
a technology of cancer immunotherapy and car therapy, which is applied in the direction of specific cell targeting fusion, antibody medical ingredients, drug compositions, etc., can solve the problems of inability to achieve routine clinical practice, inability to reduce tumor reactivity, and dire lack of antigens downregulated in tumor cells but present in normal tissue,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
REFERENCES FOR EXAMPLE 1
[0580]1. Zack T I, Schumacher S E, Carter S L, Cherniack A D, Saksena G, Tabak B, Lawrence M S, Zhsng C Z, Wala J, Mermel C H, Sougnez C, Gabriel S B, Hernandez B, Shen H, Laird P W, Getz G, Meyerson M, Beroukhim R. Pan-cancer patterns of somatic copy number alteration. Nature genetics. 2013; 45:1134-1140[0581]2. Gibson W J, Hoivik E A, Halle M K, Taylor-Weiner A, Cherniack A D, Berg A, Holst F, Zack T I, Werner H M, Staby K M, Rosenberg M, Stefansson I M, Kusonmano K, Chevalier A, Mauland K K, Trovik J, Krakstad C, Giannakis M, Hodis E, Woie K, Bjorge L, Vintermyr O K, Wala J A, Lawrence M S, Getz G, Carter S L, Beroukhim R, Salvesen H B. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nature genetics. 2016; 48:848-855[0582]3. Gerlinger M, Rowan A J, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald N Q Butler ...
example 2
REFERENCES FOR EXAMPLE 2
[0606]1. Lek M, Karczewski K J, Minikel E V, Samocha K E, Banks E, Fennell T, O'Donnell-Luria A H, Ware J S, Hill A J, Cummings B B, Tukiainen T, Birnbaum D P, Kosmicki J A, Duncan L E, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper D N, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki M I, Moonshine A L, Natarajan P, Orozco L, Peloso G M, Poplin R, Rivas M A, Ruano-Rubio V, Rose S A, Ruderfer D M, Shakir K, Stenson P D, Stevens C, Thomas B P, Tiao G, Tusie-Luna M T, Weisburd B, Won H H, Yu D, Altshuler D M, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez J C, Gabriel S B, Getz G, Glatt S J, Hultman C M, Kathiresan S, Laakso M, McCarroll S, McCarthy M I, McGovern D, McPherson R, Neale B M, Palotie A, Purcell S M, Saleheen D, Scharf J M, Sklar P, Sullivan P F, Tuomilehto J, Tsuang M T, Watkins H C, Wilson J G, Daly M J, MacArthur D G, Exome Aggregation C. Analysis of...
example 3
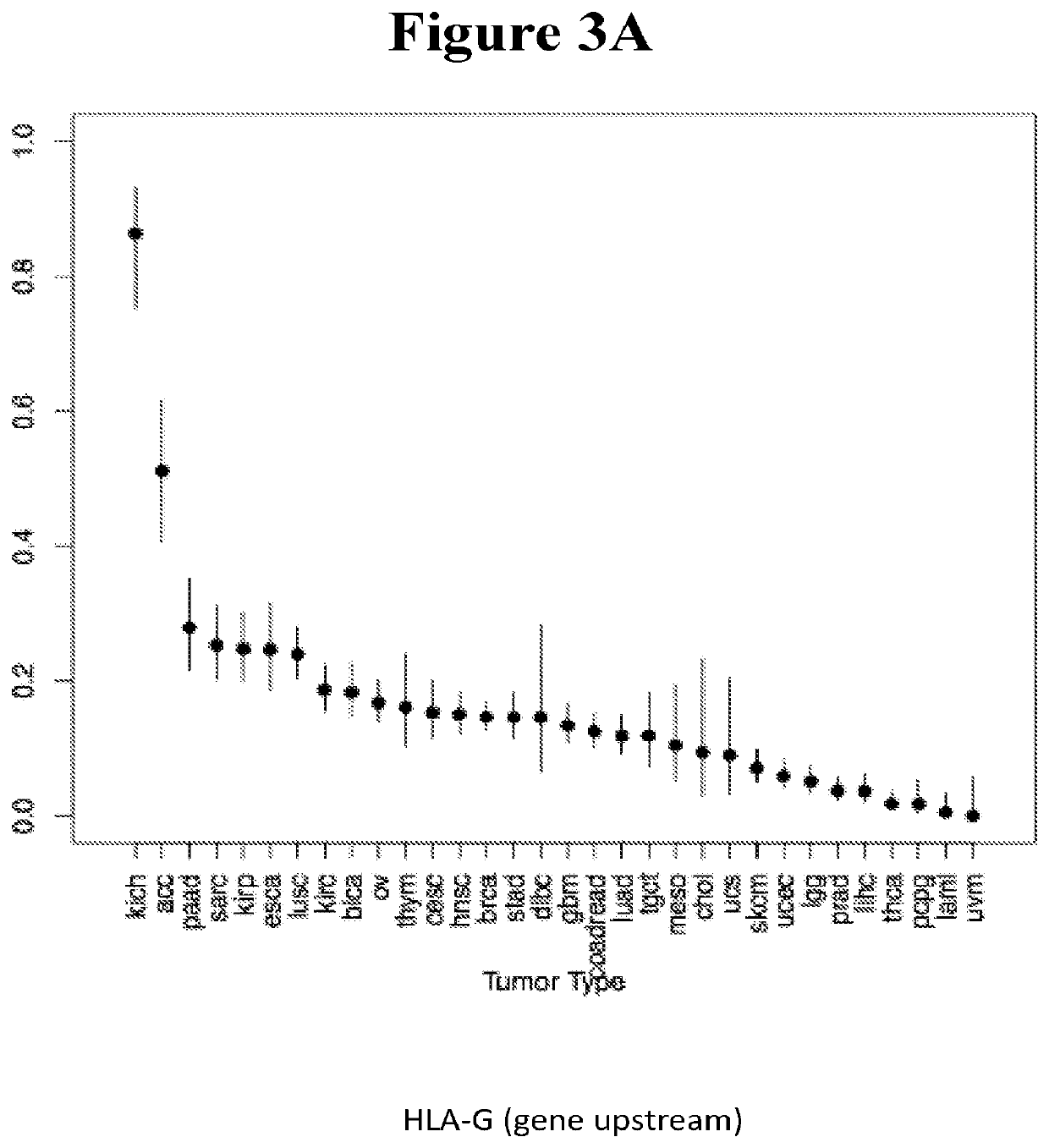
ncing Analysis for Verification of HLA LOH in Kich Samples
Library Preparation and Sequencing
[0612]Purpose—based on the in-silico analysis, KICH cancer was chosen as the first tumor type for wet verification of the HLA LOH prediction. The aim was to identify the HLA genotype for each pateinet based on DNA derived from normal tissue, and then to analyse the HLA allotype in the cancer tisse in an attempt to identify loss of one of the HLA alleles.
[0613]For that matter, HLA allotype was determined for DNA derived from 6 frozen matche KICH samples (Normal and Cancer) RC-001-RC003, TNEABAll, TNEABNWE, 2rDFRAUB, 2RDFRNQG, IOWT5AVJ, IOWT5N74. In addition, two DNA matched samples OG-001-OG-002 (Normal and Cancer) were also analysed. A DNA library was prepared sequence analysis was conducted in order to identify the sample's HLA typing. DNA was extracted from 6 frozen matched KICH samples (Normal and tumor) and a library was prepared as described below.
[0614]TruSight HLA Sequencing libraries ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| nucleic acid | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com