Identification and use of circulating nucleic acid tumor markers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Example
Example 1
An Ultrasensitive Method for Quantitating Circulating Tumor DNA with Broad Patient Coverage
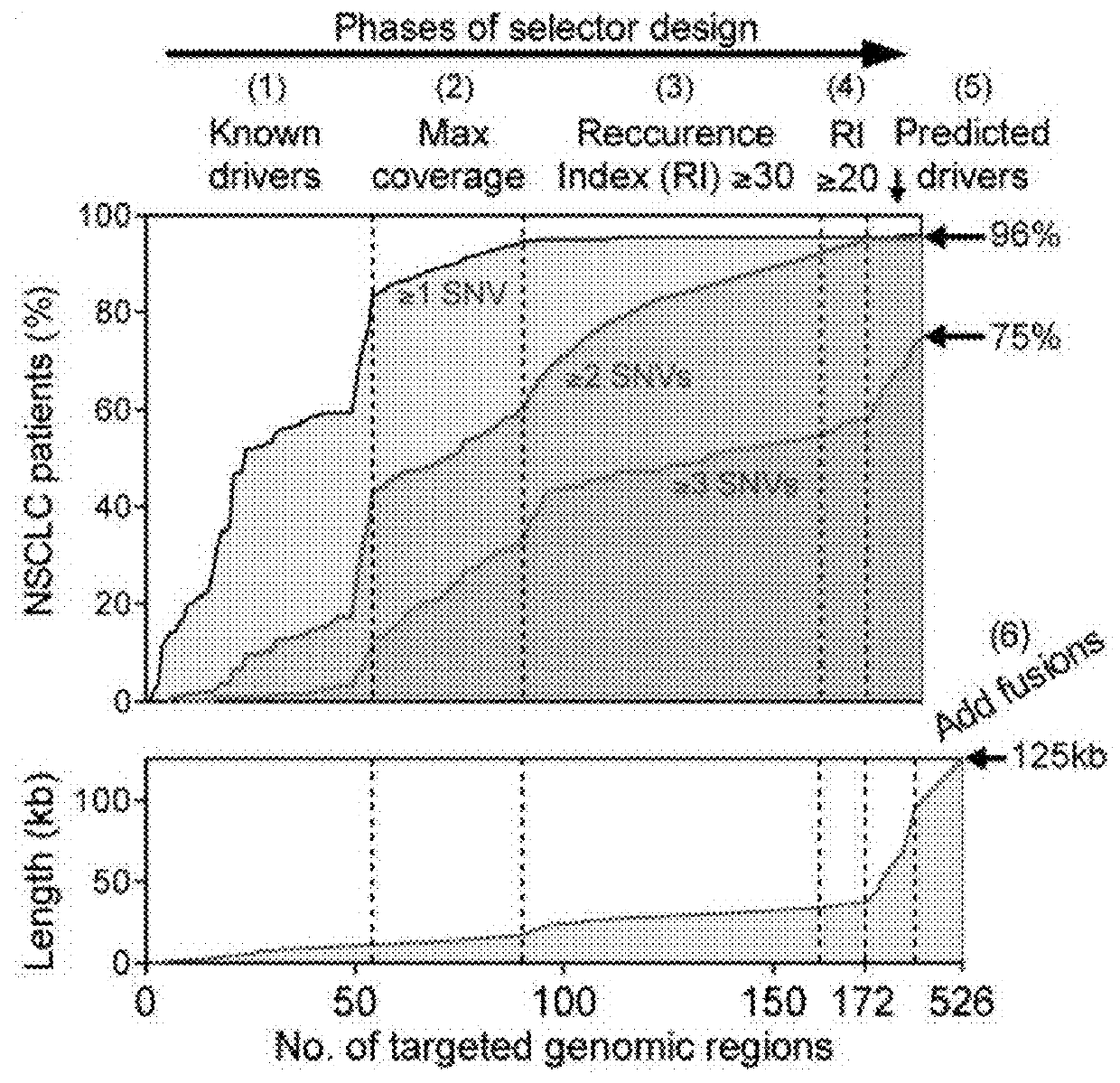
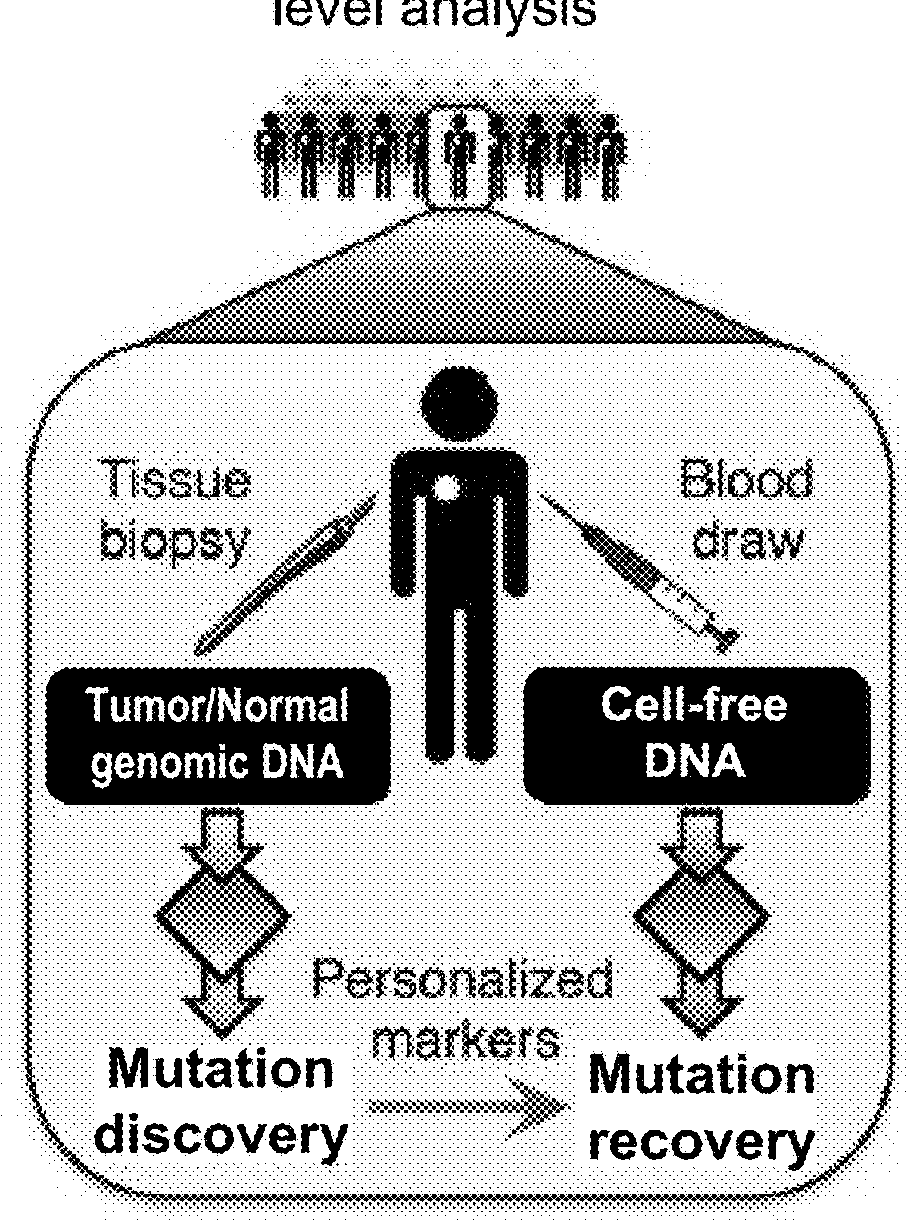
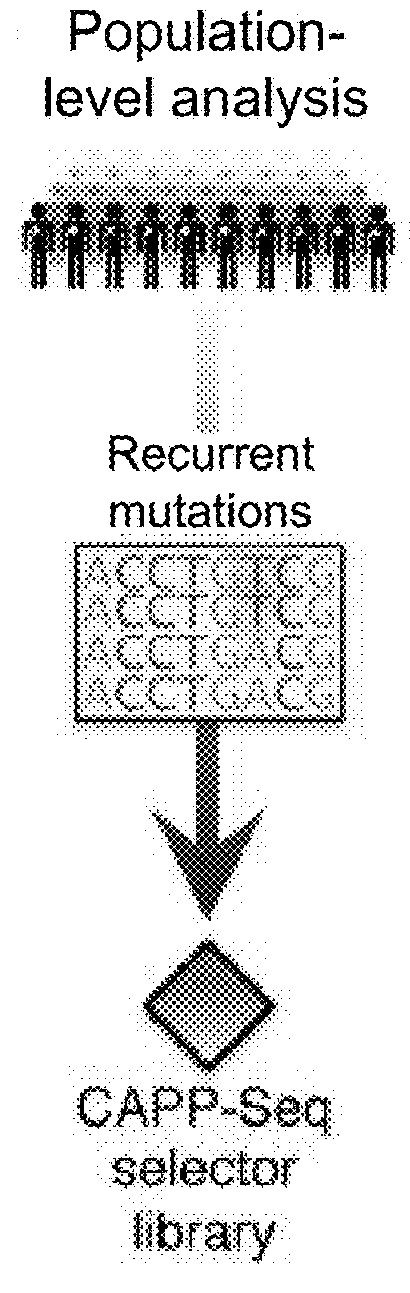
[0781]Circulating tumor DNA (ctDNA) represents a promising biomarker for noninvasive detection of disease burden and monitoring of recurrence. However, existing ctDNA detection methods are limited by sensitivity, a focus on small numbers of mutations, and / or the need for patient-specific optimization. To address these shortcomings, CAncer Personalized Profiling by Deep Sequencing (CAPP-Seq) was developed, an economical and highly sensitive method for quantifying ctDNA in plasma in nearly every patient. We implemented CAPP-Seq for non-small cell lung cancer (NSCLC) with a design that identified mutations in >95% of tumors, simultaneously detecting point mutations, insertions / deletions, copy number variants, and rearrangements. When tumor mutation profiles were known, we detected ctDNA in 100% of pre-treatment plasma samples from stages II-IV NSCLC and 50% of samples from stage I NSCLC,...
Example
Example 2
Designing a Personalized Selector Set
[0879]In certain circumstances, monitoring tumor burden in a patient known to have cancer is likely to be impractical using an ‘off-the-shelf’ strategy applying knowledge from a cohort of patients with the same tumor type, to selectively capture genomic regions that are recurrently mutated in that tumor type using CAPP-Seq. These situations include, but are not limited to, cases where (1) the tumor is of an unknown primary histology (e.g., CUP); (2) the histology is known, but is too rare to have a sufficient number of patients with that tumor type previously profiled to define the average patient's tumor somatic genetic landscape (e.g., soft tissue sarcoma subtyped); (3) the histology is known but the average / median number of recurrent somatic lesions in that tumor type are too low to achieve desired sensitivity levels (e.g., pediatric tumors, etc.); or (4) the histology is known and the average / median number of recurrent somatic lesion...
Example
Example 3
Use of a Selector Set to Diagnose a Cancer
[0881]A plasma sample is obtained from a female subject with an abnormal lump in her breast. Cell-free DNA (cfDNA) is extracted from the plasma sample. An end repair reaction is performed on the cfDNA by mixing the components in a sterile microfuge tube (or other suitable sterile container) as follows:
ComponentVolume (μL)cfDNA1-75Phosphorylation Reaction Buffer (10X)10T4 DNA polymerase5T4 Polynucleotide kinase5dNTPs4DNA Polymerase I, Large (Klenow)1Sterile H2O-bring total volume up to 100 μL
[0882]The end repair reaction mixture is incubated in a thermal cycler for 30 minutes at 20° C.
[0883]Clean-up of the end repaired cfDNA is performed by adding 160 μL (1.6×) of resuspended AMPure XP beads to the end repair reaction mixture. The AMPure beads are mixed into the solution on a vortex mixer or by pipetting up and down (e.g., 10 times or more). The reaction is incubated for 5 minutes at room temperature. The reaction is placed on a magn...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Length | aaaaa | aaaaa |
| Length | aaaaa | aaaaa |
| Length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com