Prokaryote molecule label method based on RNA polymerase binding site
An RNA polymerase, prokaryotic technology, applied in the field of prokaryotic molecular labeling, can solve the problems of low annealing temperature and false positives, and achieve the effects of good repeatability, high specificity and high stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
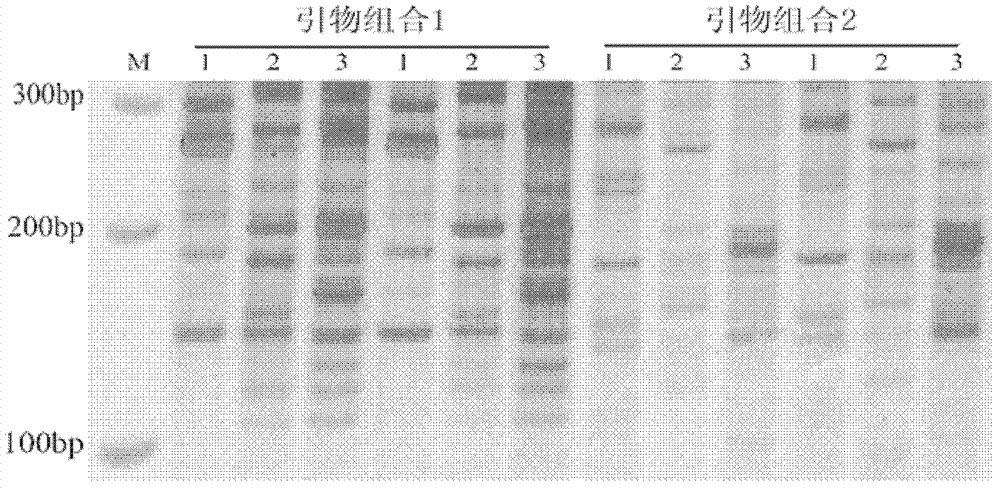
[0017] The present invention will be described in detail below in conjunction with embodiments and drawings, but the present invention is not limited to this.
[0018] A method for labeling prokaryotic molecules based on RNA polymerase binding sites includes the following steps:
[0019] (1) Design primers
[0020] Design a set of forward primers: 5’GACTGCGTACGTTGACAN 3’ (N is any of the 4 bases A / T / G / C);
[0021] A set of reverse primers: 5'GACTGCGTACGTATANNN 3'(N is any one of the 4 bases of A / T / G / C).
[0022] (2) Synthetic primer
[0023] Send the primer sequence to Shanghai Shenggong Biological Engineering Co., Ltd. for synthesis. After obtaining the primer, it will be prepared to the required concentration and stored at -20°C for later use.
[0024] (3) DNA extraction
[0025] The rhizosphere soil of three plants of rice, broad bean and lettuce were collected, and the total DNA of rhizosphere soil microorganisms was extracted by CTAB method as a template. The DNA quality and concentr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com