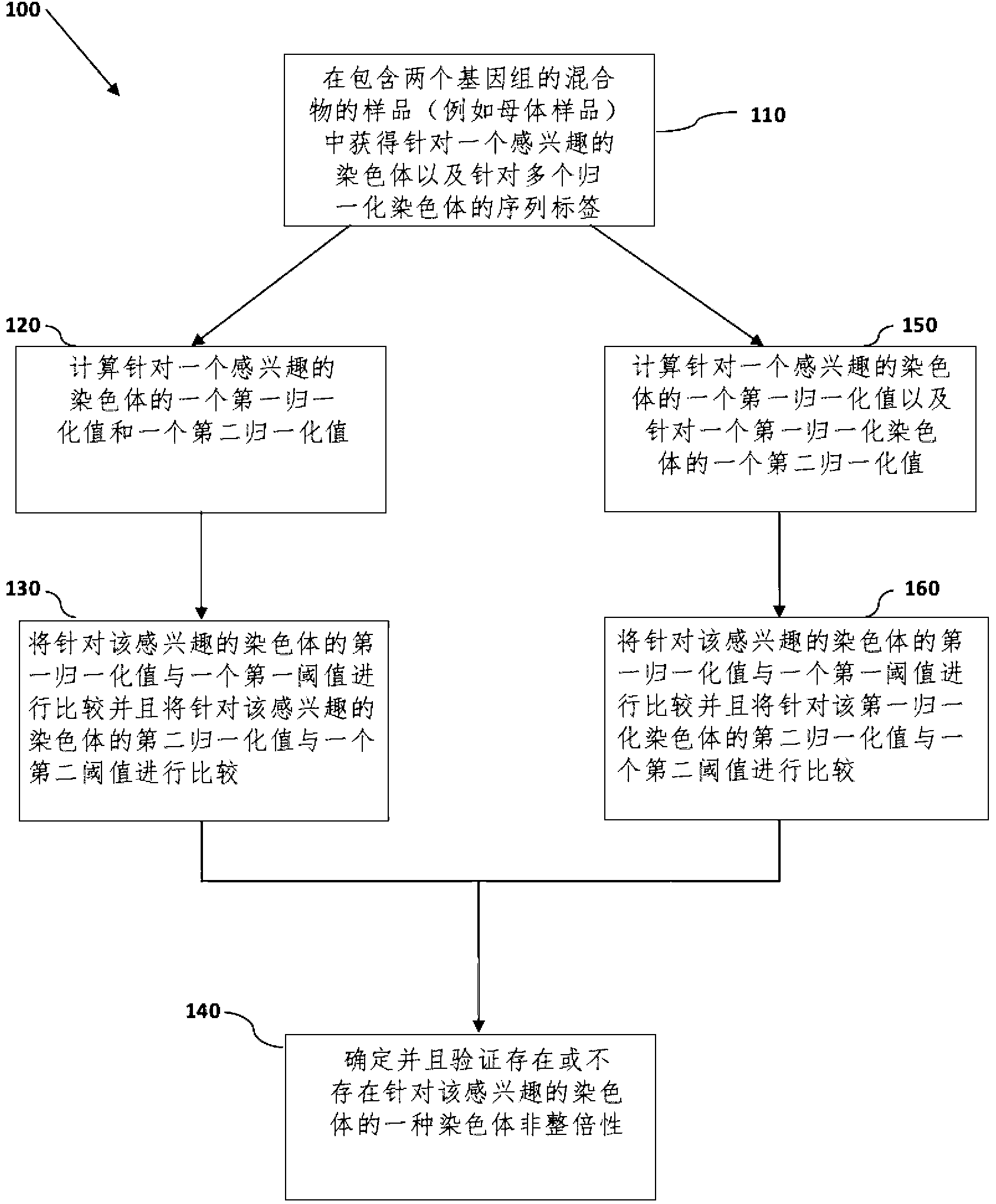
Normalizing chromosomes for the determination and verification of common and rare chromosomal aneuploidies
A technique for aneuploidy and chromosomes, applied in the determination/inspection of microorganisms, special data processing applications, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0186] Optimal determination of fetal chromosomal abnormalities using massively parallel DNA sequencing of cell-free fetal DNA from maternal blood: a test set 1 independent of the training set 1
[0187] This study was conducted by qualified site clinical investigators at 13 U.S. clinical sites between April 2009 and October 2010 as a scientific trial in human subjects approved by each institution's Institutional Review Board (IRB) planned. Written consent was obtained from each subject prior to participation in the study. The scientific trial program is designed to provide blood samples as well as clinical data to support the development of noninvasive prenatal genetic diagnostic methods. Pregnant women 18 years of age or older are eligible to participate. Blood was collected from patients undergoing clinically indicated chorionic villus sampling (CVS) or amniocentesis prior to the procedure, and results of fetal karyotype were also collected. Peripheral blood samples (two...
example 2
[0248] Validating aneuploidy determinations using multiple chromosome ratios: normalizing normalizing chromosomes
[0249] As described in previous examples, the method is based on the number of sequence tags mapped to the chromosome of interest for sequence tags mapped to samples exhibiting similar inter-sample and inter-run variability to the chromosome of interest The normalization of the number of . In order to validate the classification of aneuploidy and rule out that the normalizing chromosomes used in the analysis are themselves aneuploid chromosomes (i.e. present in abnormal copy numbers), the first normalizing chromosomes (i.e. used to determine chromosome dosage in Normalization for chromosomes) used to classify common aneuploidies involving chromosomes 21, 18, and X.
[0250] Using qualifying samples from training set 1 as described in Example 1, and qualifying samples from test set 1, the sequencing information was analyzed to identify at least one second normali...
example 3
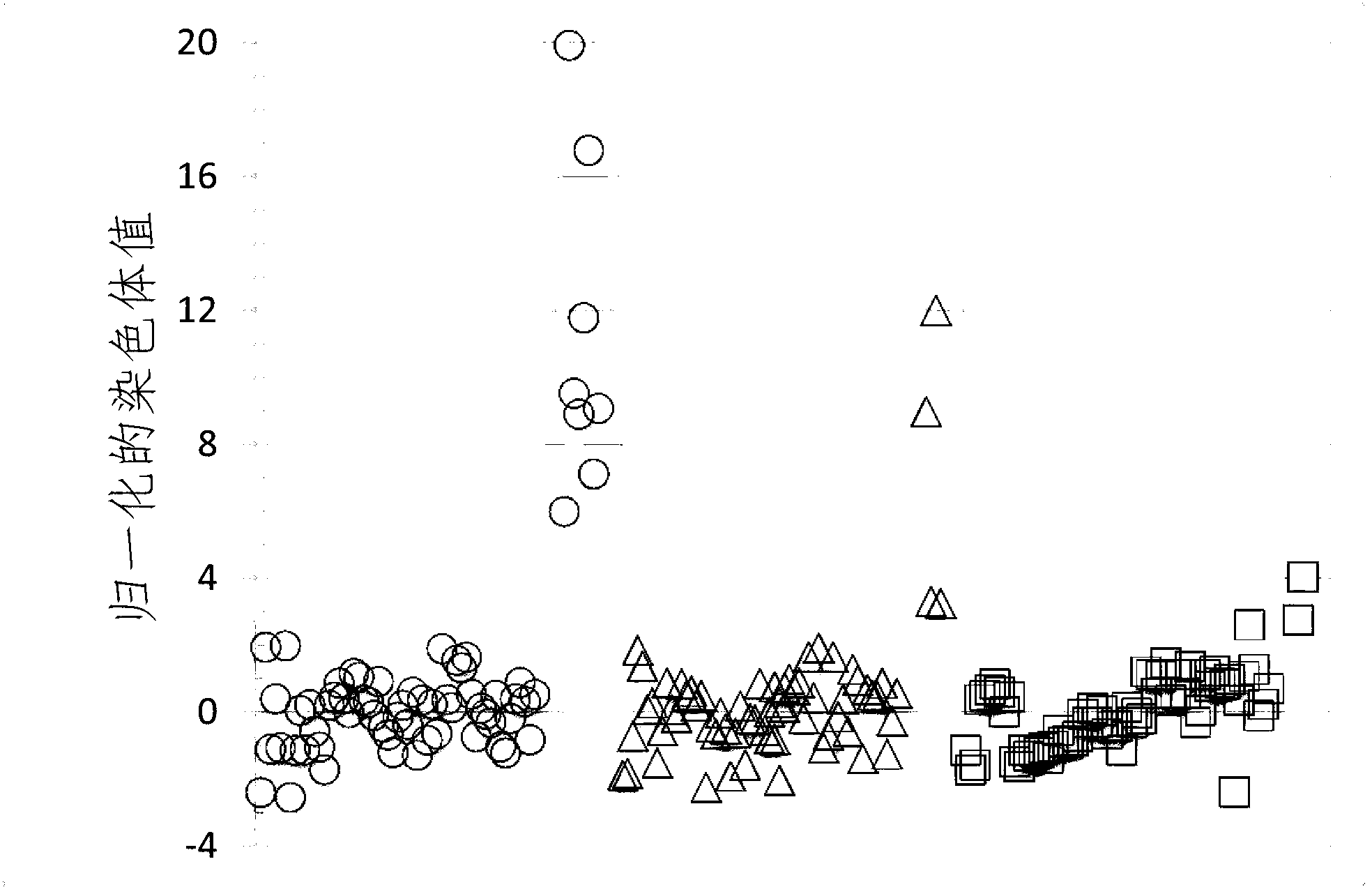
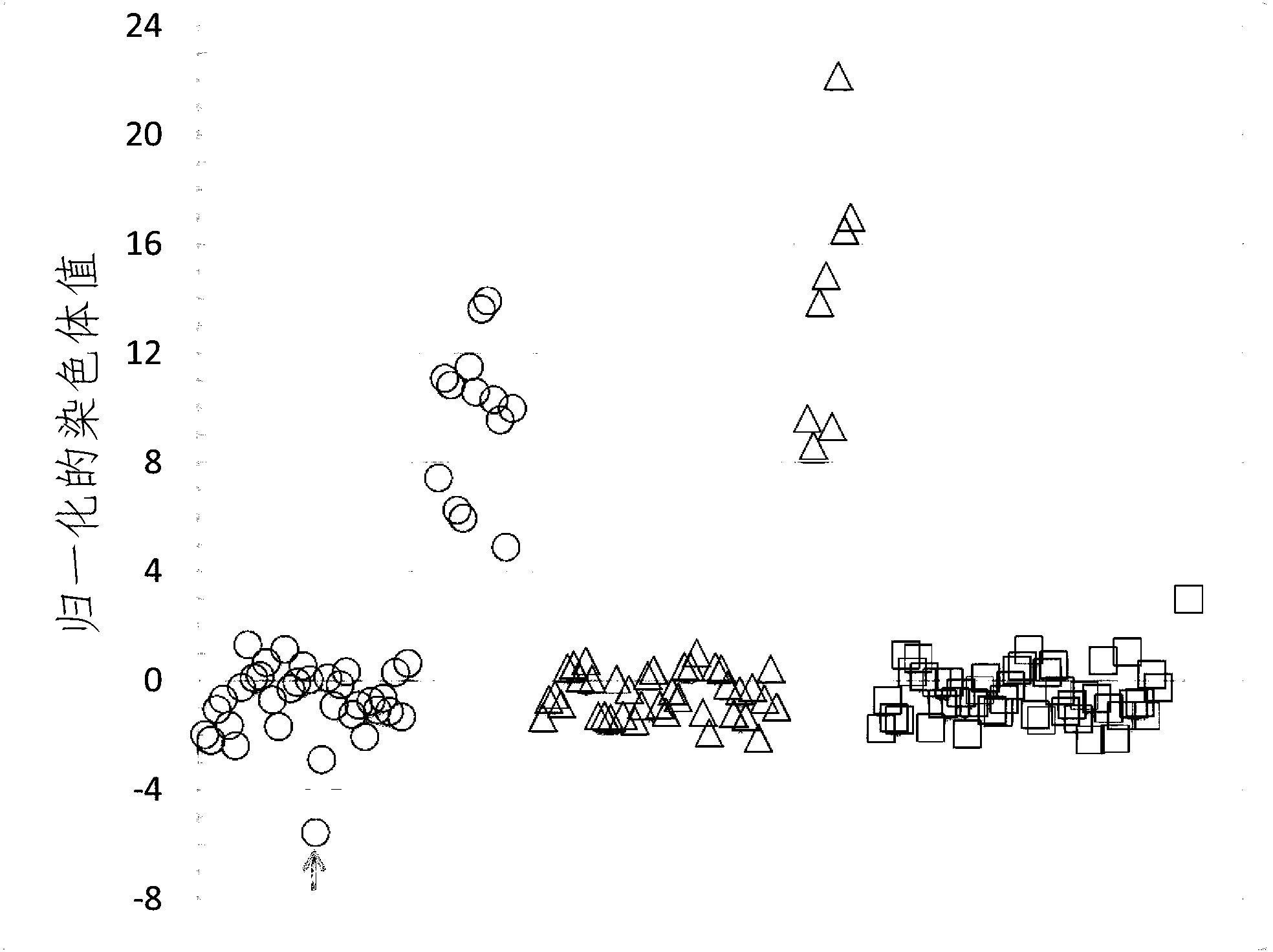
[0279] Determine and verify chromosomal aneuploidy using at least two normalizing chromosomes for the chromosome of interest
[0280] To demonstrate that the determination of chromosomal aneuploidy can be validated by using the first and second normalizing chromosomes against the chromosome of interest, using chromosome 10 and chromosome 14 as the second and third normalizing against chromosome 21 of interest Normalizing Chromosome, Chromosome Dose for Chromosome 21 in Example 1A calculated using Chromosome 9 as the first Normalizing Chromosome.
[0281] FIG. 8A shows a plot of the NCVs for the 48 samples in test set 1 calculated using the mean and S.D. of the corresponding chromosome doses in the qualifying samples in training set 1 . The average CV % of chromosome dosage for chromosome 21 in training set 1 is provided in Table 7.
[0282] Table 7
[0283] Determination of the second normalizing chromosome for the chromosome of interest chromosome 21
[0284]
[0285] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com