Method and product for localised or spatial detection of nucleic acid in a tissue sample
A tissue sample, local detection technology, which is applied in the preparation of test samples, biochemical equipment and methods, and the determination/inspection of microorganisms, etc., can solve the problem of no research on high-throughput methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0359] Array preparation
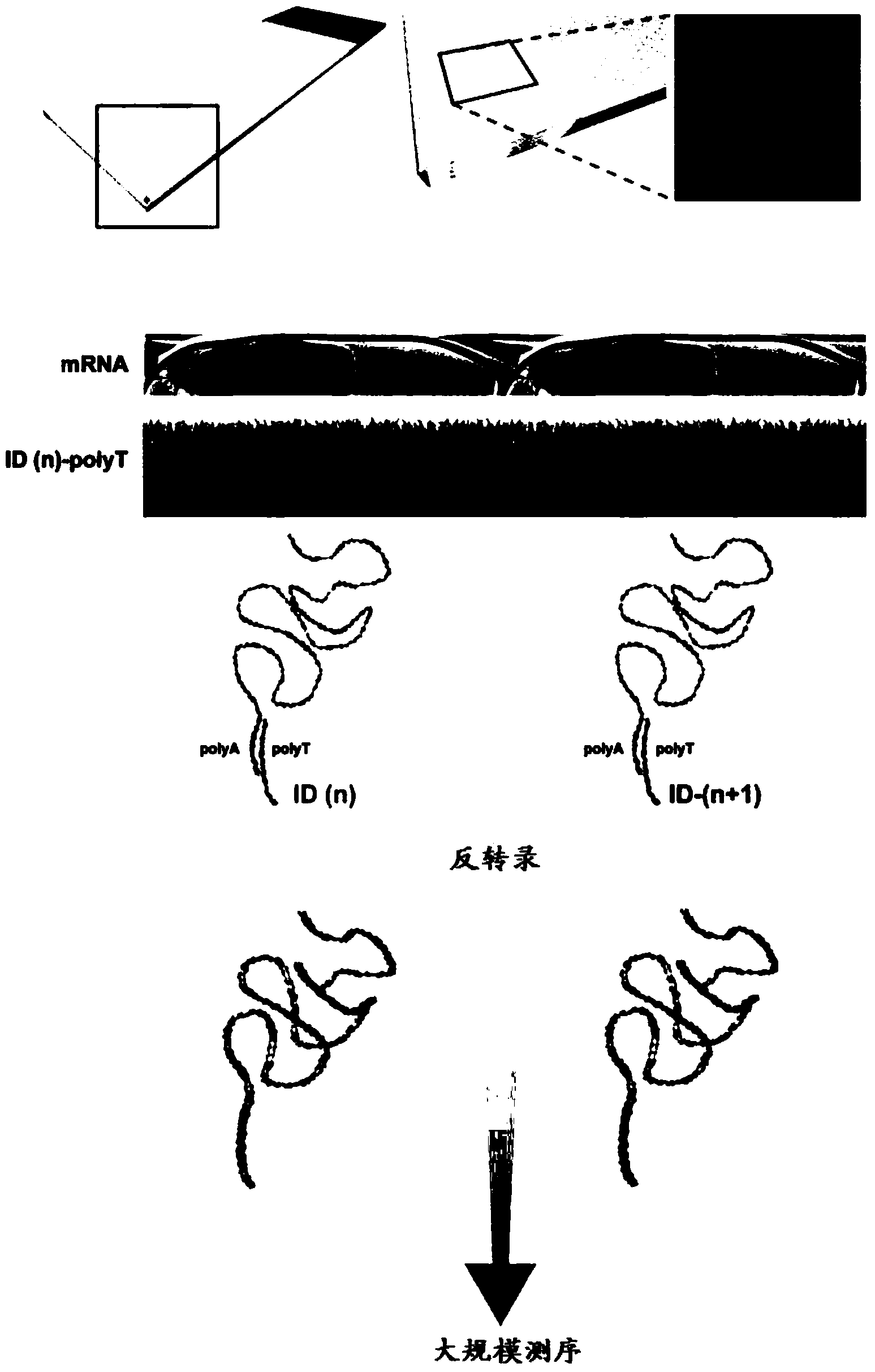
[0360] The following experiments demonstrate how to attach oligonucleotide probes to array substrates via their 5' or 3' ends to obtain arrays containing capture probes that hybridize to mRNA.
[0361] Preparation of self-made spray-dot microarray with probes in 5' to 3' direction
[0362] Twenty RNA capture oligonucleotides (labeled 1-20, Table 1) each with a labeled sequence were spotted on a glass slide as capture probes. The probe was synthesized with a 5'-terminal amino linker and a C6 spacer. All probes were synthesized by Sigma-Aldrich (St. Louis, MO, USA). The RNA capture probe was prepared with 150 mM sodium phosphate to make a suspension with a concentration of 20 μM and a pH of 8.5, and the Nanoplotter NP2.1 / E spraying system (Gesim, Grosserkmannsdorf, Germany) was used in CodeLink TM Activated microarray slides (7.5cm x 2.5cm; Surmodics, Eden Prairie, MN, USA) were spotted. After spraying is complete, follow the manufacturer's inst...
Embodiment 2
[0433] Figure 8 to Figure 12 Demonstrates successful visualization of stained FFPE mouse brain tissue (olfactory bulb) sections placed on transcriptome capture arrays labeled with identifiers, following the general protocol described in the Examples. Compared with the experiments of fresh frozen tissue in Example 1, Figure 8 The FFPE tissue morphology shown is better. Figure 9 and Figure 10 Shows how to localize tissue on arrays of different probe density types.
Embodiment 3
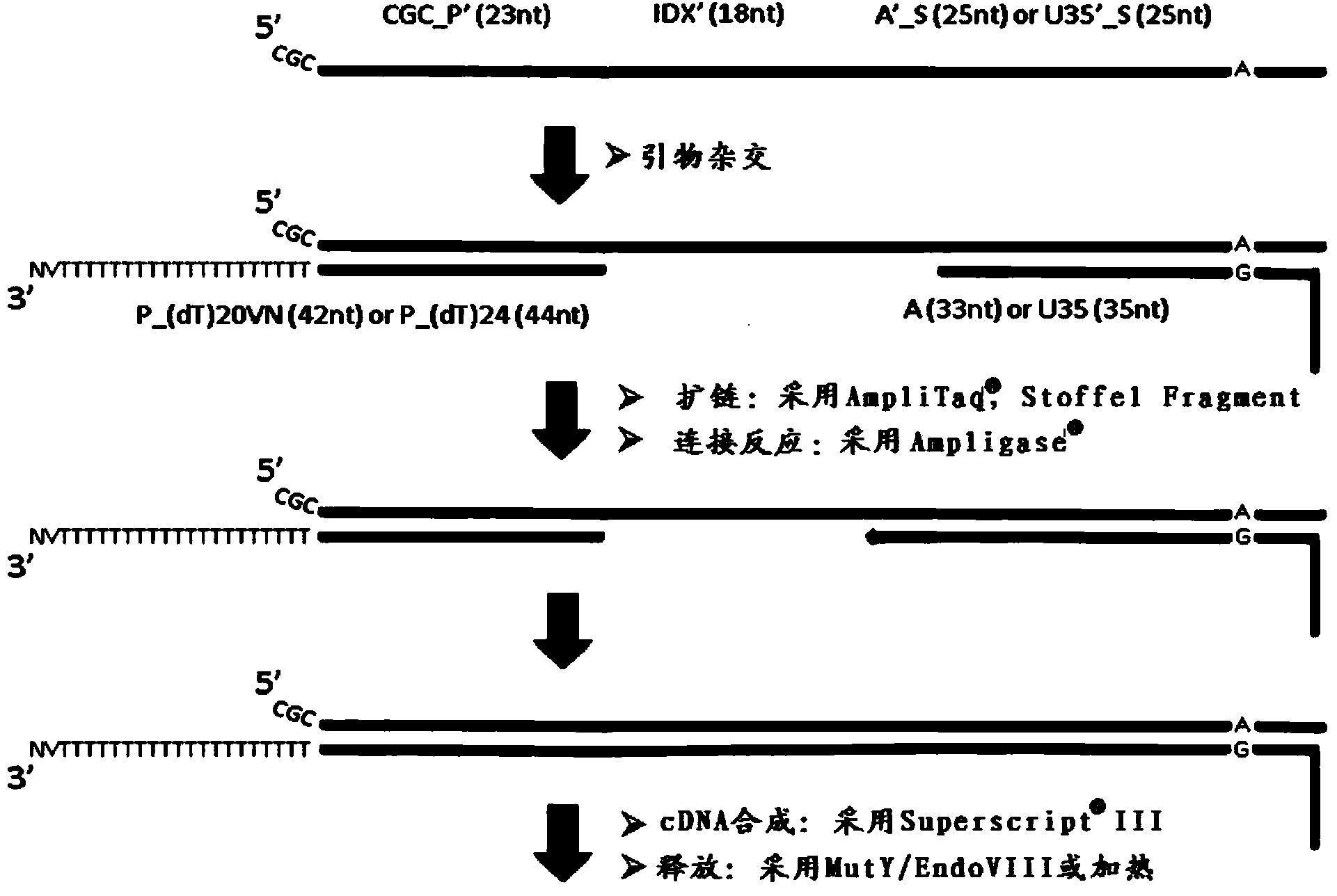
[0435] Whole-transcriptome amplification with random-primed second-strand synthesis and universal-end amplification (capture probes containing tag sequences retain at the end of the resulting dsDNA)
[0436] After release of capture probes with PCR buffer containing USER enzyme mix capable of cleaving uracil (covalently attached probes)
[0437] or
[0438] After release of capture probes with heated PCR buffer (synthetic capture probes for in situ hybridization)
[0439] Add 40μl of 1x containing 1.8mM MgCl to each of the 2 tubes 2 Fast start high-fidelity PCR buffer (Faststart HiFi PCR Buffer with1.8mM MgCl 2 ) (pH8.3) (Roche, www.roche-applied-science.com), dNTP 0.2mM each (Fermentas, www.fermentas.com), 0.1μg / μl BSA (New England Biolabs, www.neb.com) , 0.1 U / μl USER enzyme (New England Biolabs), released cDNA (extended from surface probe) and released surface probe, and 1 μl RNaseH (5 U / μl). Then, 2 tubes were incubated in a thermal cycler (Applied Biosystems, www....
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap