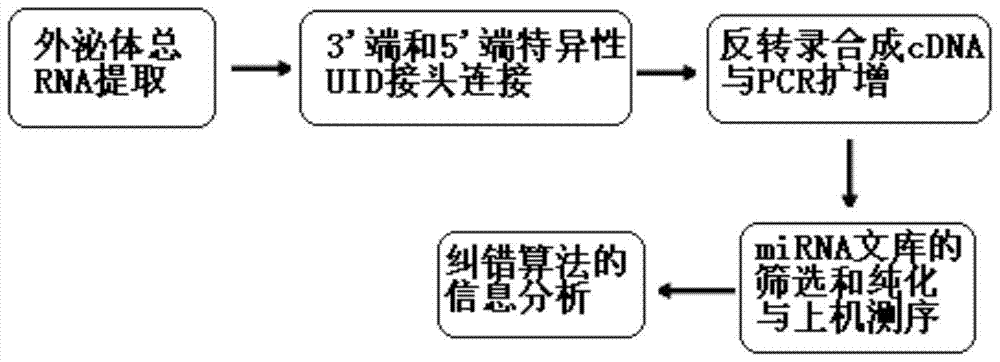
A quantitative detection method for exosome miRNA
A quantitative detection and exosome technology, applied in the field of quantitative detection of exosome miRNA, can solve the problems of high error rate of preference, influence on detection, and few miRNAs, and achieve the effect of correcting related sequencing errors and avoiding preference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Example 1 Establishment of Exosome miRNA Quantitative Detection Method
[0062] 1. Extraction of total RNA from exosomes:
[0063] 1.1 Draw 2 tubes (5mL / tube) of peripheral blood from the subject into EDTA anticoagulant tubes, gently upside down (to prevent cell rupture), mix thoroughly 6-8 times, and perform the following treatments within 4-6 hours on the day of blood collection; Centrifuge at 1600g for 10 minutes at 4°C. After centrifugation, divide the supernatant (plasma) into multiple 1.5mL / 2mL centrifuge tubes. During the suction process, the white blood cells in the middle layer cannot be absorbed; centrifuge at 16,000g for 10 minutes at 4°C. Minutes, remove the residual cells, transfer the supernatant (plasma) to a new 1.5mL / 2mL centrifuge tube, if the white blood cells cannot be sucked to the bottom of the tube, the required plasma after separation is obtained;
[0064] 1.2 After the plasma sample was processed, all the separated plasma (about 5ml) was extrac...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com