A gene mutation detection method and reagent for reducing amplification bias
A detection method, a technology to be detected, applied in the direction of biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of low yield of amplification products, low-abundance DNA mutations are difficult to achieve sufficient copy number, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
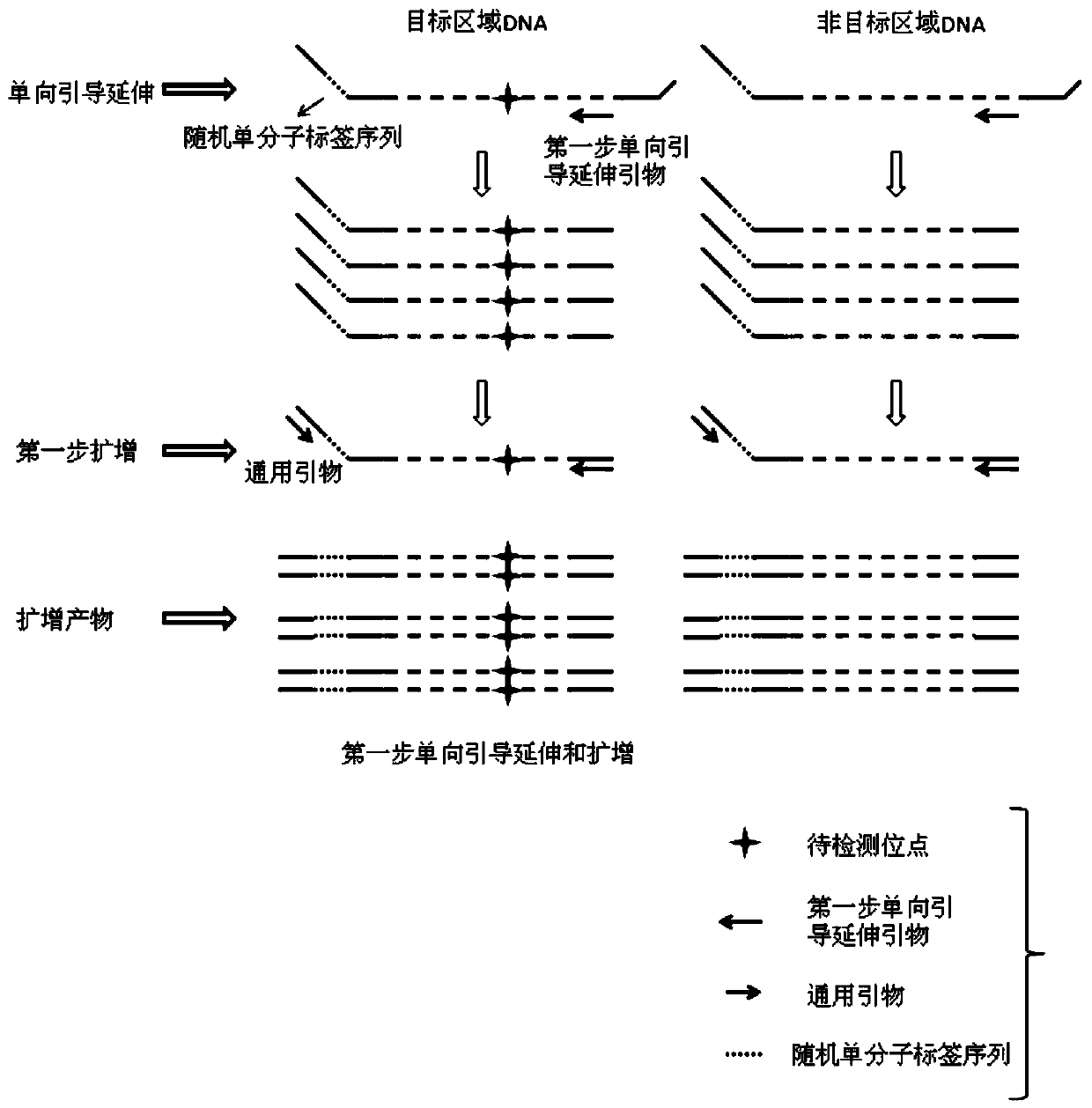
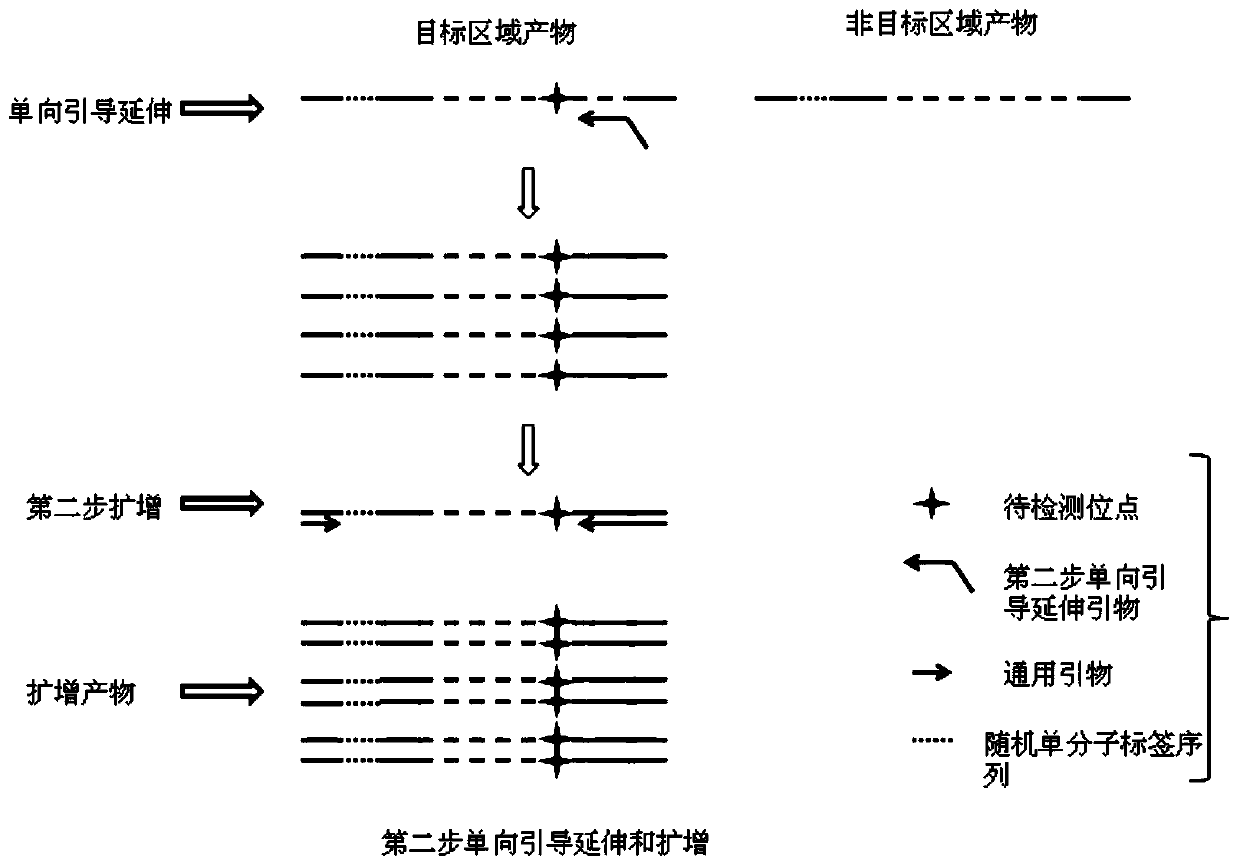
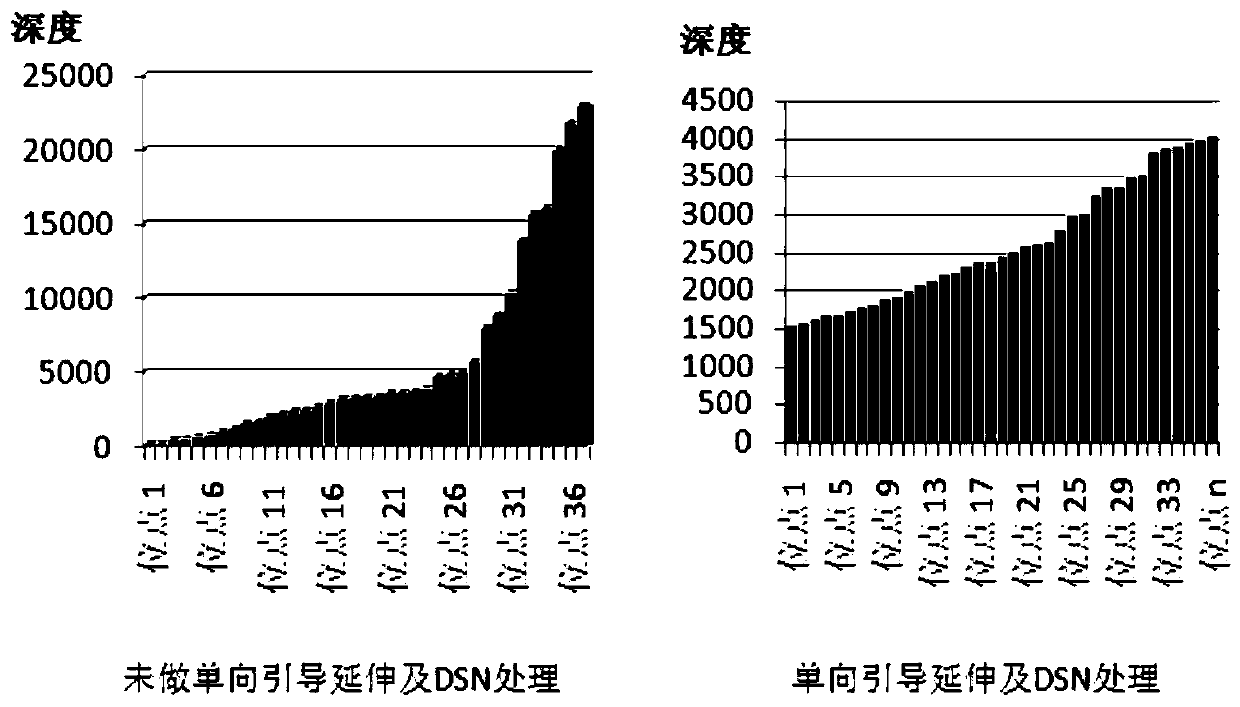
[0060] In this example, library construction was performed on samples and seven sites of 4 genes NRAS, KRAS, PI3KA, and EGFR were detected, as follows:
[0061] 1. Adapter Design Containing Random Single Molecular Tags
[0062] IDX1-S:
[0063] CAAGCAGAAGACGGCATACGAGATNNNNNNNggaattaGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT (SEQ ID NO: 1);
[0064] IDX2-S:
[0065] CAAGCAGAAGACGGCATACGAGATNNNNNNNNatccggcGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT (SEQ ID NO: 2);
[0066] IDX3-S:
[0067] CAAGCAGAAGACGGCATACGAGATNNNNNNNNNcaggccgGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT (SEQ ID NO: 3);
[0068] ADT-AS: pGATCGGAAGAGC (SEQ ID NO: 4); the phosphate group at the 5' end of the ADT-AS sequence is modified.
[0069] IDX1-S, IDX2-S, and IDX3-S (both are the first strands of the linker) are annealed with the ADT-AS sequence respectively to form double strands, constituting the three linkers ADT1, ADT2, and ADT3 of the present invention.
[0070] 2. This example detects seven common mutation points in the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com