Rapid single cell library building method
A single-cell, rapid technology, applied in the fields of biochemical equipment and methods, chemical libraries, combinatorial chemistry, etc., can solve the problems of increasing sequencing sample preparation time and reagent cost, masking, easy contamination, etc., and achieve extensive sequencing coverage, The effect of shortening sequencing time and reducing amplification bias
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
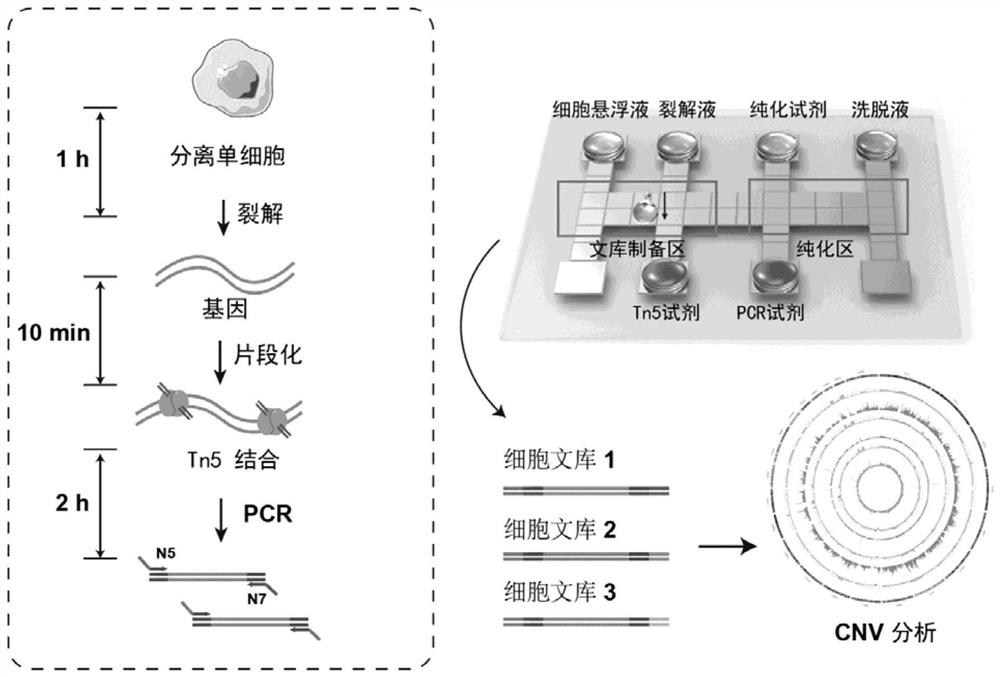
[0056] The adherent human embryonic lung fibroblast MRC-5 (purchased from the Cell Bank of the Type Culture Collection Committee of the Chinese Academy of Sciences, catalog number GNHu41) was used as the model cell for the experiment.
[0058] Use the capillary pipetting technique to obtain MRC-5 single cells, place them in a PCR tube, add 1 μL of lysate, and place the PCR tube on a PCR instrument to heat and lyse. The lysate contains 60mM Tris-HCl pH 8.0, 2mM EDTA, 15mM DTT and 0.5mg / ml Protease K, and the lysis program is 50°C for 1h, 80°C for 10min.
[0059] 2. Genome Fragmentation
[0060] use DNA Library Prep Kit V2for (TD503-01 / 02) kit, prepare Tn5 transposase mixture, as shown in Table 1. Add 2 μL of Tn5 transposase mixture to PCR, wherein, 5×TBBL is Tn5 transposase buffer, TTE Mix: Tn5 transposase. Run the program on the PCR instrument: 55°C, 10min.
[0061] Table 1. Tn5 Transposase Mixture Components
[0062] Element ...
Embodiment 2
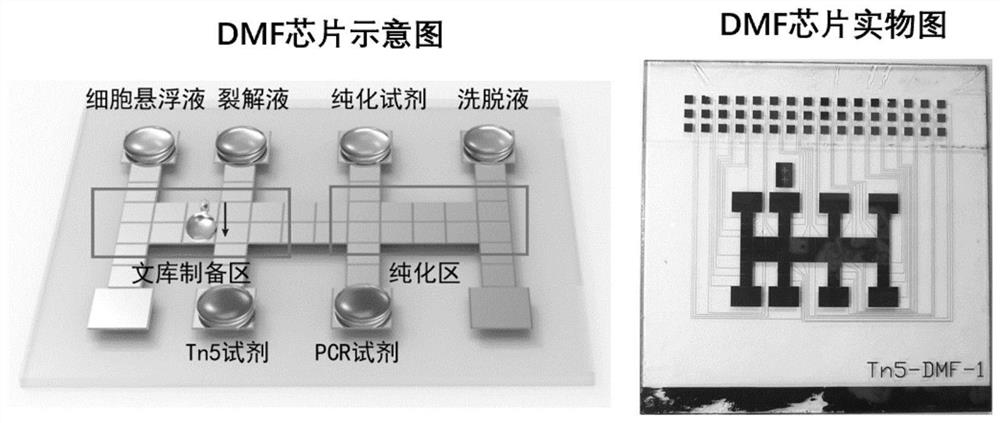
[0089]This embodiment includes isolation of single cells, single cell lysis, genome fragmentation and whole genome amplification reactions. Specifically, first use the digital microfluidic chip to separate single cells; then add cell lysate to lyse single cells; then add Tn5 transposase mixture to use Tn5 transposase to fragment DNA; finally add barcodes and fragments containing different indexes The PCR Mix of the stop solution was used for PCR amplification. Amplified products can be directly purified and sequenced.
[0090] During the experiment, the adherent cell MRC-5 was used as the model cell for direct single-cell bank building. The specific experimental steps and results are as follows:
[0091] 1. DMF chip design and production:
[0092] In this embodiment, a digital microfluidic chip known in the art can be used, for example, refer to the digital microfluidic chip shown in Ruan et al., Sci.Adv.2020; 6:eabd6454; Anal.Chem.2020,92,8599-8606 Fluidic chip.
[0093]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com