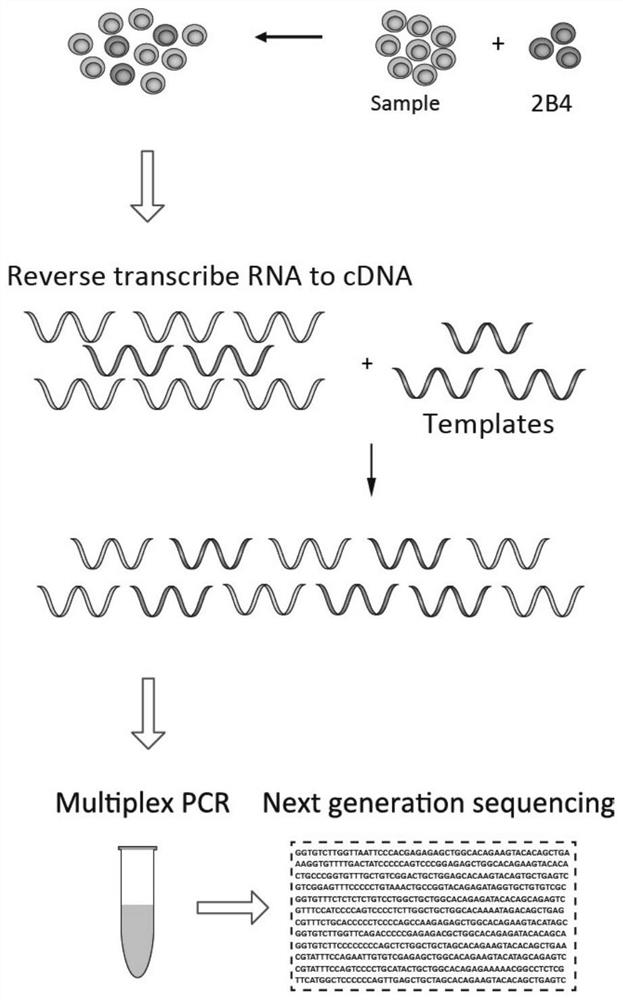
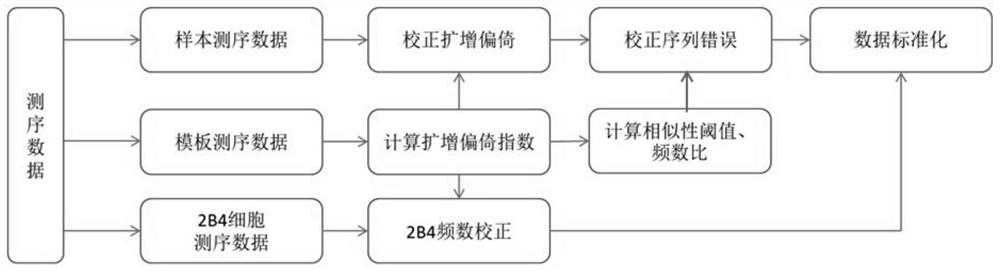
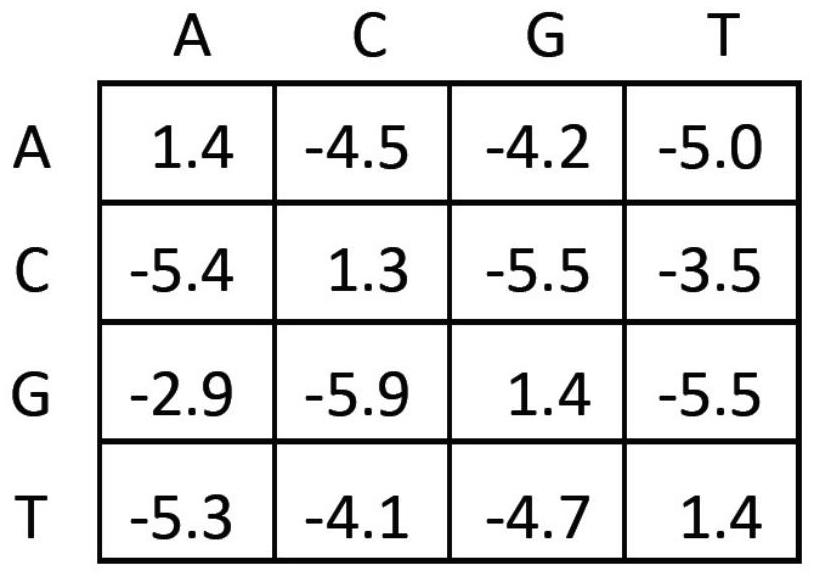
Method for quantifying TCR beta based on high-throughput sequencing
A high-throughput, sequencing technology, applied in biochemical equipment and methods, microbial measurement/inspection, sequence analysis, etc., can solve problems such as sequencing data amplification bias, sequencing errors, and the estimated impact of T cell pool diversity , to reduce interference and amplification bias
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] TCR β library construction and sequencing of mouse spleen CD3+ T cells.
[0058] 1. RNA extraction
[0059] Sorting 1,000,000 mouse spleen CD3+ T cells, adding 800ul Trizol (TRIzol Reagent, Invitrogen, 15596018), pipetting and mixing, standing at room temperature for 5min, adding 200ul Trizol solution dissolved with 200 2B4 hybridoma cells; adding 200ul of chloroform, mix by inversion for 30s, and place at room temperature for 3min; centrifuge at 12000g for 15min at 4°C; absorb the upper aqueous phase, and transfer it to another EP tube; 10min; 4°C, centrifuge at 12000g for 10min; add 75% ethanol to 1ml 75% ethanol / ml Trizol, shake gently, suspend the sediment; 4°C, 8000g centrifuge for 5min, suck off the supernatant; Dissolve in RNase-free water to obtain total mouse RNA, and perform concentration determination and quality control.
[0060] The extracted RNA was used to measure the RNA concentration and quality by epoch. The test results showed that OD260 / OD...
Embodiment 2
[0095] Quantitative analysis of TCR β in mouse spleen CD3+ T cells.
[0096] The TCR number measured by evaluating the external reference cells is used as a reference for the number of T cells in the sample; the sequencing error is corrected by the template sequence. Utilizing the assumption that "high-frequency sequences are more likely to be the original correct sequences", the sequence errors are corrected using the Stepwise extraction clustering method. Use the molecular barcode in the template sequence to separate the template sequence from the sequencing sample, count the number of sequencing reads of different V template sequences, investigate the law of the amplification bias after the sample is mixed, explore the method of correcting the amplification bias, and correct Sequence errors generated during sequencing.
[0097] This example is based on the mouse spleen CD3+ T cell sequencing data (see Example 1) to study the characteristics of the TCR β pool in the m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com