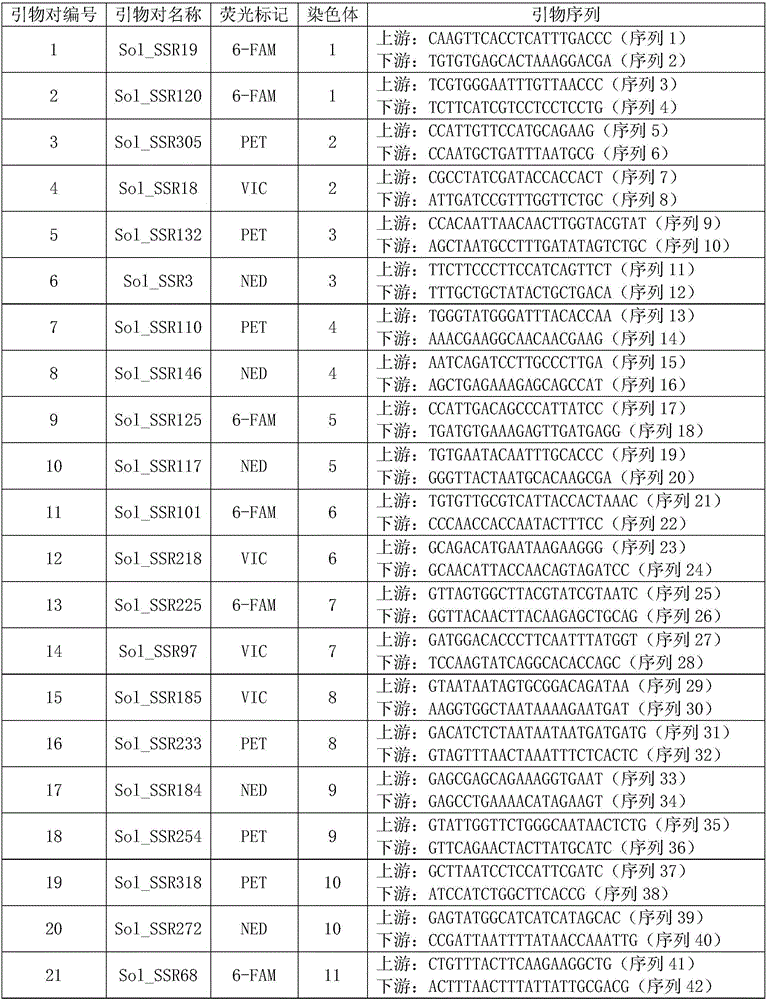
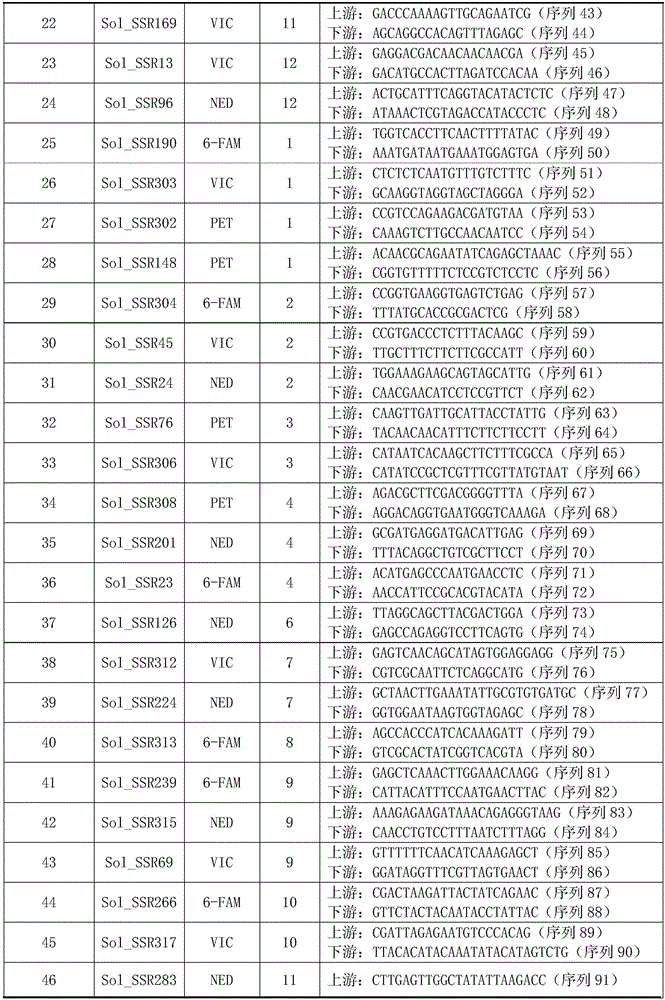
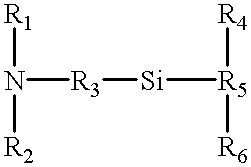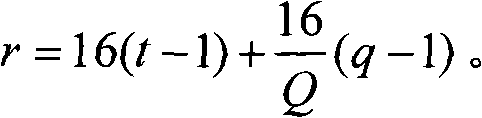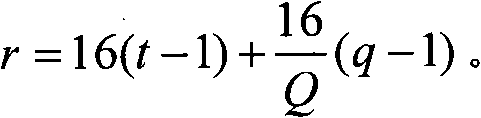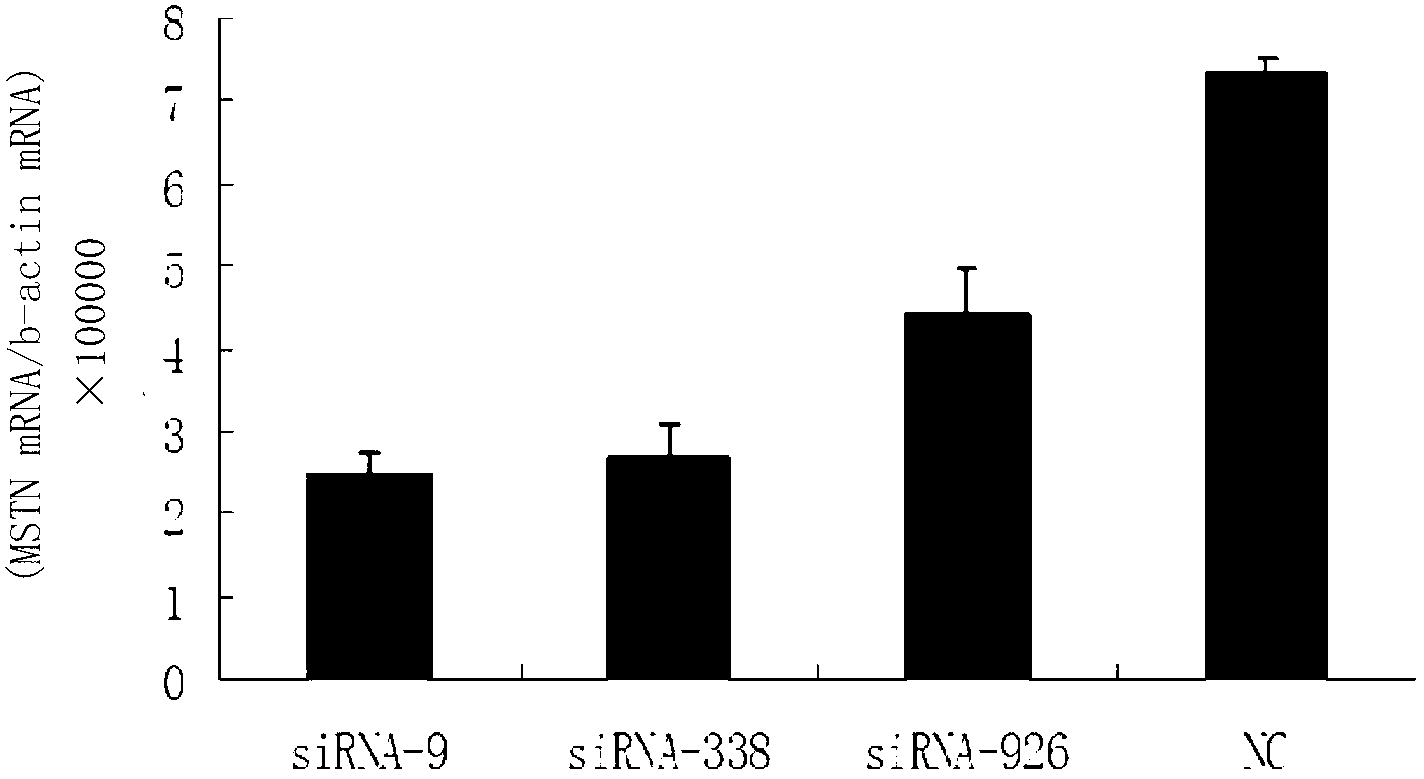Patents
Literature
212 results about "Sequenceome" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Sequenceome is the totality of polymer sequences on Earth. Sequences in biology such as genome and proteome are the major components. Sequenceome is effectively a large database.
Methods and devices for measuring differential gene expression
InactiveUS6355423B1Efficient and accurateImprove search efficiencySugar derivativesMicrobiological testing/measurementSequence databaseNucleic Acid Sequence Databases
This invention includes methods for identifying nucleic acids in a sample of nucleic acids by observing sequence sets present in the nucleic acids of the sample and then identifying those sequences in a nucleic acid sequence database having the sequence sets observed. In a preferred embodiment, a sequence set consists of two primary subsequences and an additional subsequence having determined mutual relationships. The methods include those for observing the sequence sets and those for performing sequence database searches. This invention also includes devices for recognizing in parallel the additional subsequences in a sample of as well as methods for the use of these devices. In a preferred embodiment, the devices include probes bound to a planar surface that recognize additional subsequence by hybridization, and the methods of use include features to improve the specificity and reproducibility of this hybridization.
Owner:CURAGEN CORP
Methods and apparatus for high-speed approximate sub-string searches
InactiveUS6931401B2Data processing applicationsMicrobiological testing/measurementSequence searchSequence database
A method and system for conducting sequence searches in a sequence database wherein in one embodiment, the method includes: combining a plurality of query sequences into a combined query sequence; determining a plurality of subdivisions of the sequence database; performing a plurality of searches, wherein each search includes a comparison of the combined query sequence against one of the plurality of subdivisions of the database to produce a plurality of word matches; extending the length of the plurality of word matches to produce a plurality of High-scoring Segment Pairs, combining the plurality of High-scoring Segment Pairs; and producing a plurality of reports, each report representing the highest scoring matches for one of the plurality of query sequences.
Owner:PARACEL
Methods and devices for measuring differential gene expression
InactiveUS20030003463A1Efficient and accurateImprove search efficiencyMicrobiological testing/measurementBiological testingSequence databaseNucleic Acid Sequence Databases
This invention includes methods for identifying nucleic acids in a sample of nucleic acids by observing sequence sets present in the nucleic acids of the sample and then identifying those sequences in a nucleic acid sequence database having the sequence sets observed. In a preferred embodiment, a sequence set consists of two primary subsequences and an additional subsequence having determined mutual relationships. The methods include those for observing the sequence sets and those for performing sequence database searches. This invention also includes devices for recognizing in parallel the additional subsequences in a sample of as well as methods for the use of these devices. In a preferred embodiment, the devices include probes bound to a planar surface that recognize additional subsequence by hybridization, and the methods of use include features to improve the specificity and reproducibility of this hybridization.
Owner:CURAGEN CORP
Methods and compositions to detect nucleic acids in a biological sample
InactiveUS8034554B2Bioreactor/fermenter combinationsBiological substance pretreatmentsNucleotideOrganic chemistry
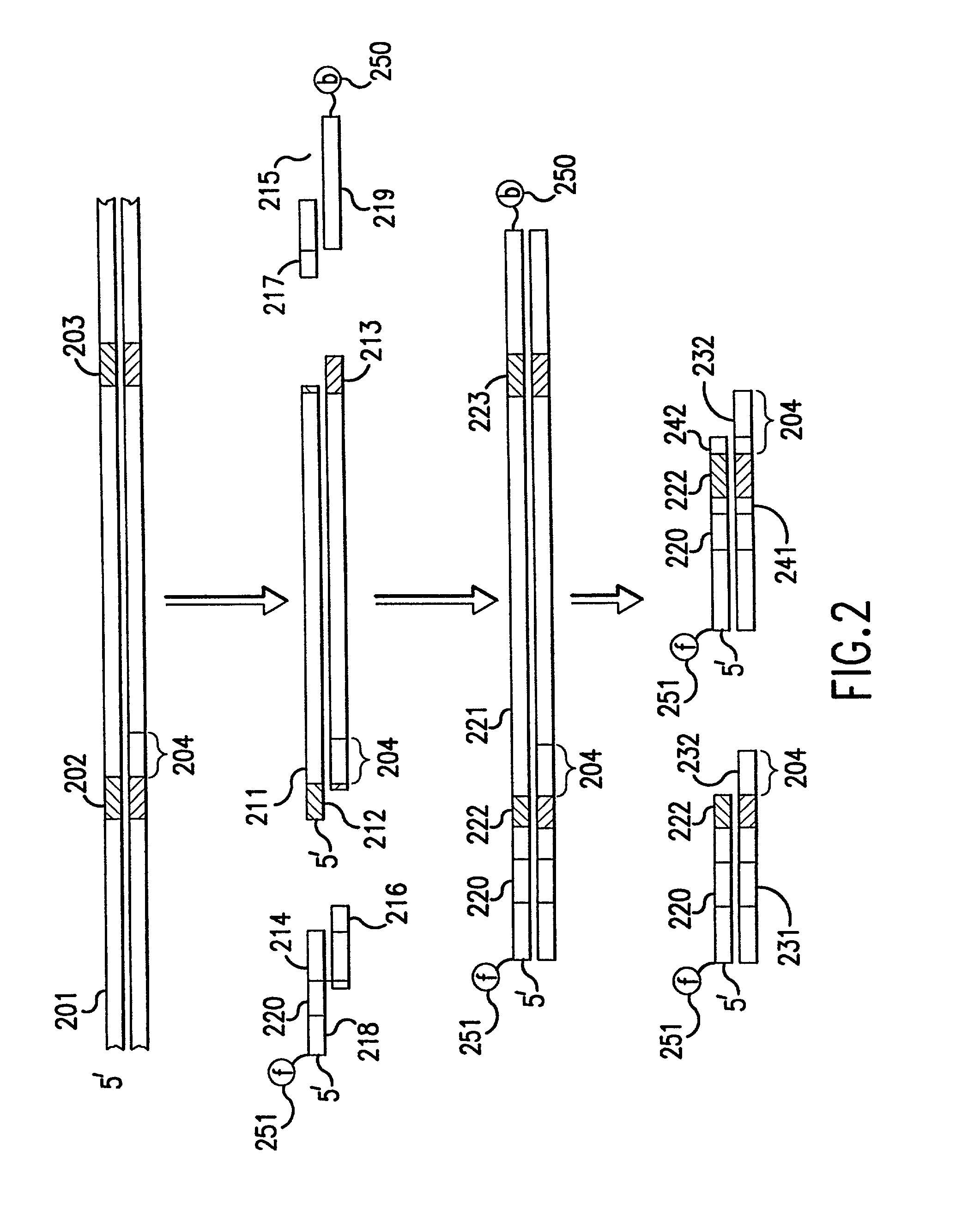
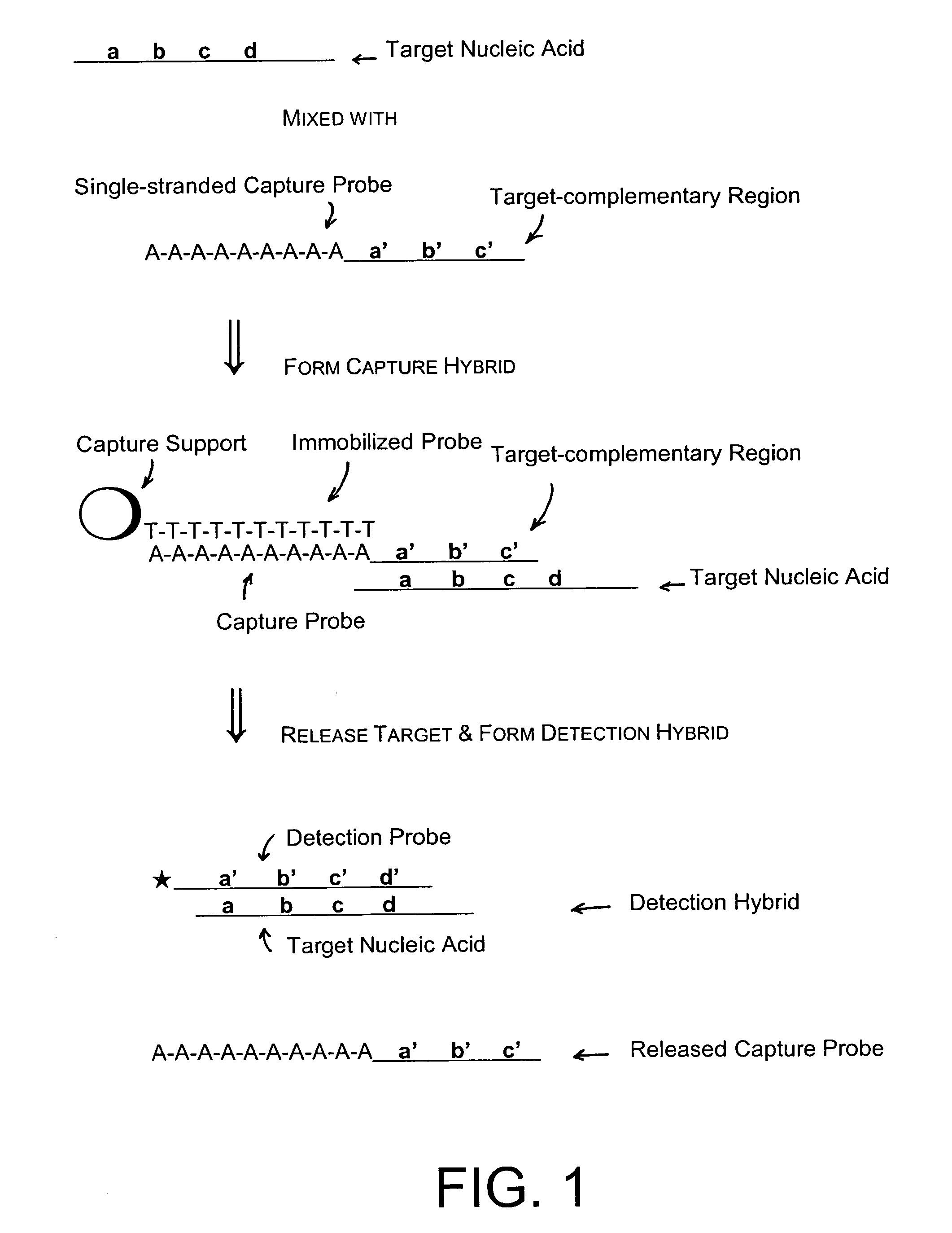
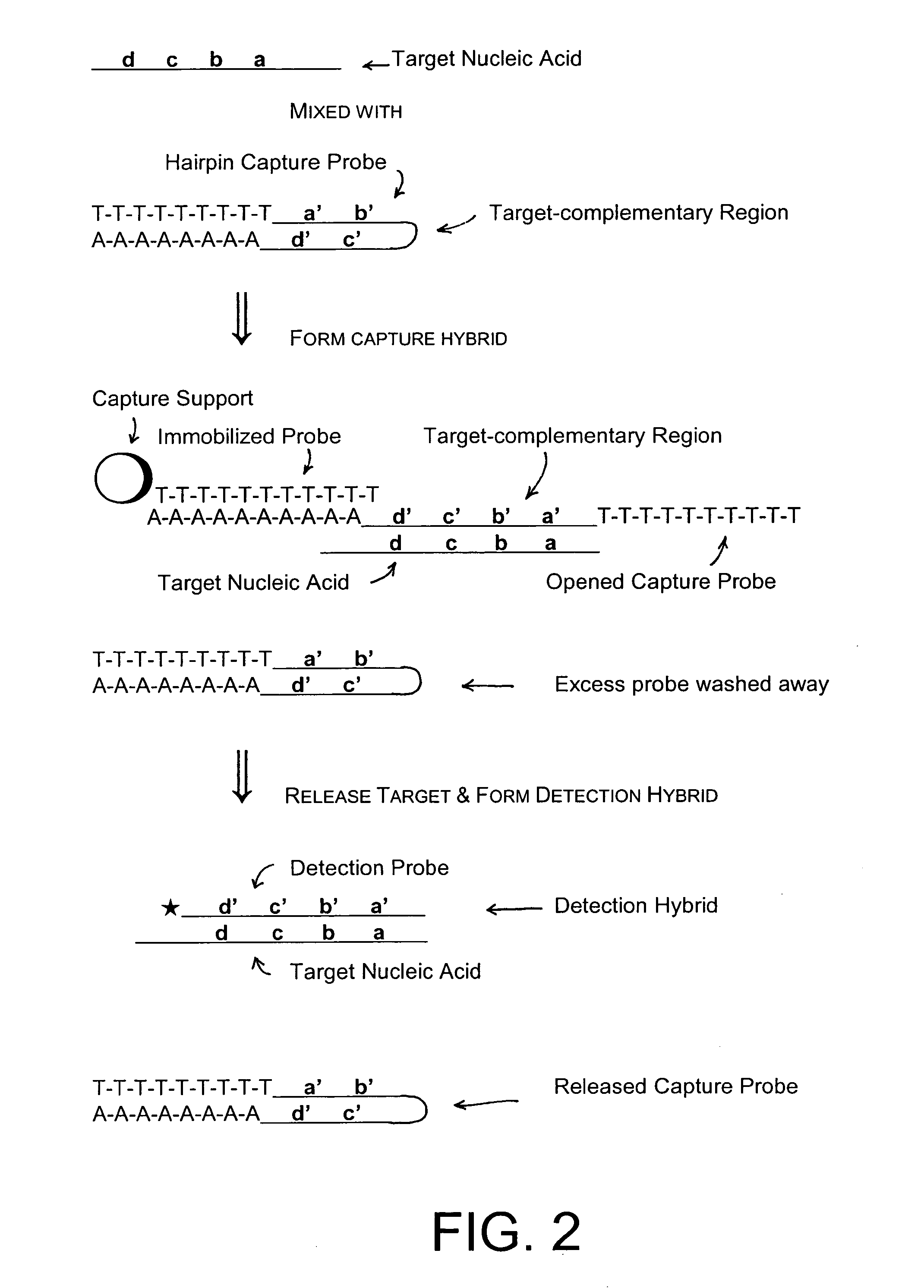
Methods of the invention separate a target nucleic acid from a sample by using at least one capture probe oligonucleotide that contains a target-complementary region and a member of a specific binding pair that attaches the target nucleic acid to an immobilized probe on a capture support, thus forming a capture hybrid that is separated from other sample components before the target nucleic acid is released from the capture support and hybridized to a detection probe to form a detection hybrid that produces a detectable signal that indicates the presence of the target nucleic acid in the sample. Compositions for practicing the methods of the invention include a capture probe oligonucleotide made up a target-complementary region sequence and a covalently linked capture region sequence that includes a member of a specific binding pair.
Owner:GEN PROBE INC
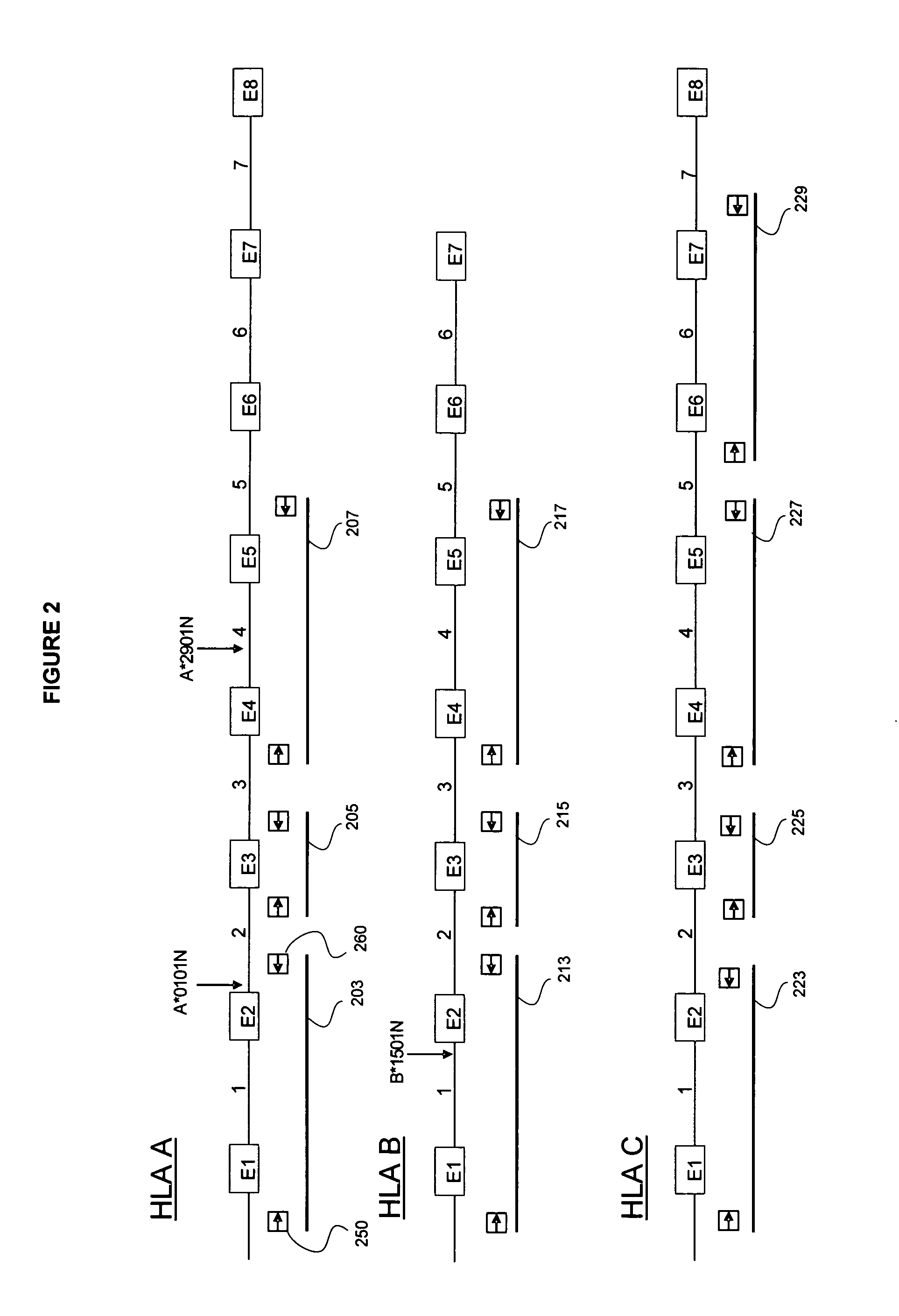
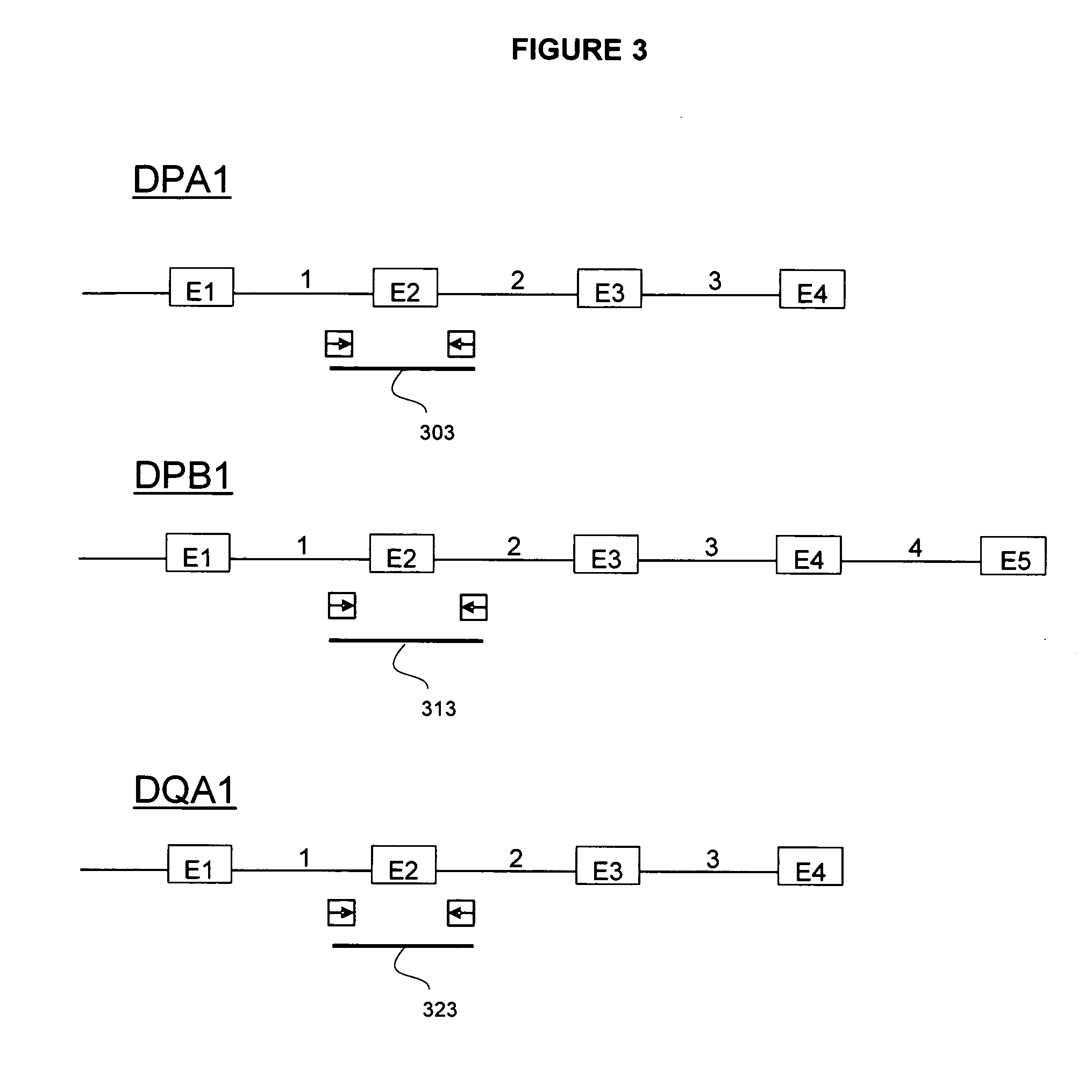
System and method for detection of HLA Variants
A method for detecting one or more HLA sequence types is described that comprises the steps of: amplifying a plurality of first amplicons from a double stranded nucleic acid sample, wherein the first amplicons are amplified with a plurality of pairs of nucleic acid primers that define exons 2 and 3 of both strands of HLA loci from the group consisting of HLA-A, HLA-B, and HLA-C; amplifying the first amplicons to produce a plurality of populations of second amplicons, wherein each population of second amplicons is clonally amplified from one of the first amplicons; sequencing the plurality of populations of second amplicons to generate a nucleic acid sequence composition for each of the plurality of second amplicons; and detecting variation in the sequence composition from one or more of the second amplicons for one or more of the HLA loci.
Owner:454 LIFE SCIENCES CORP
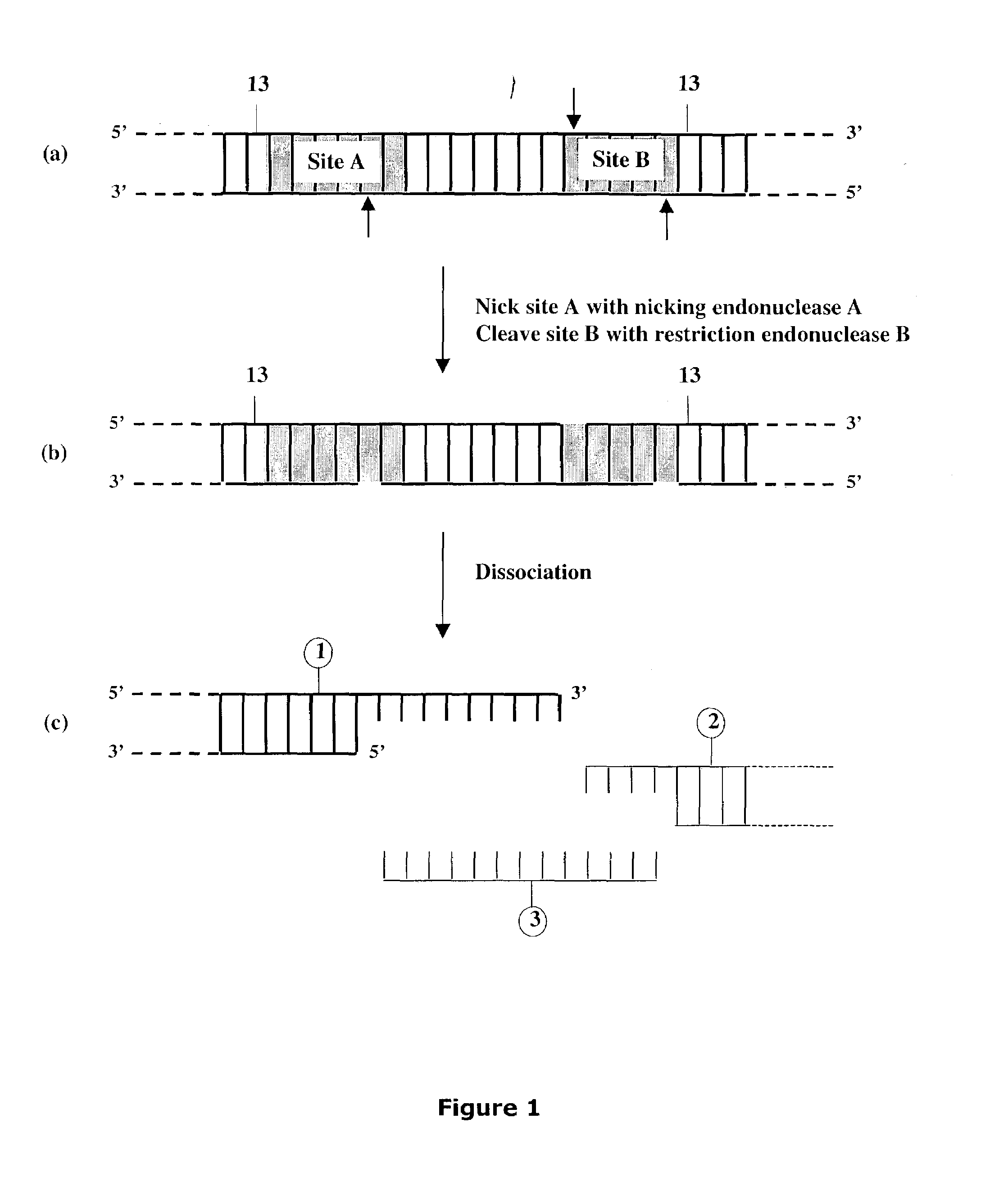
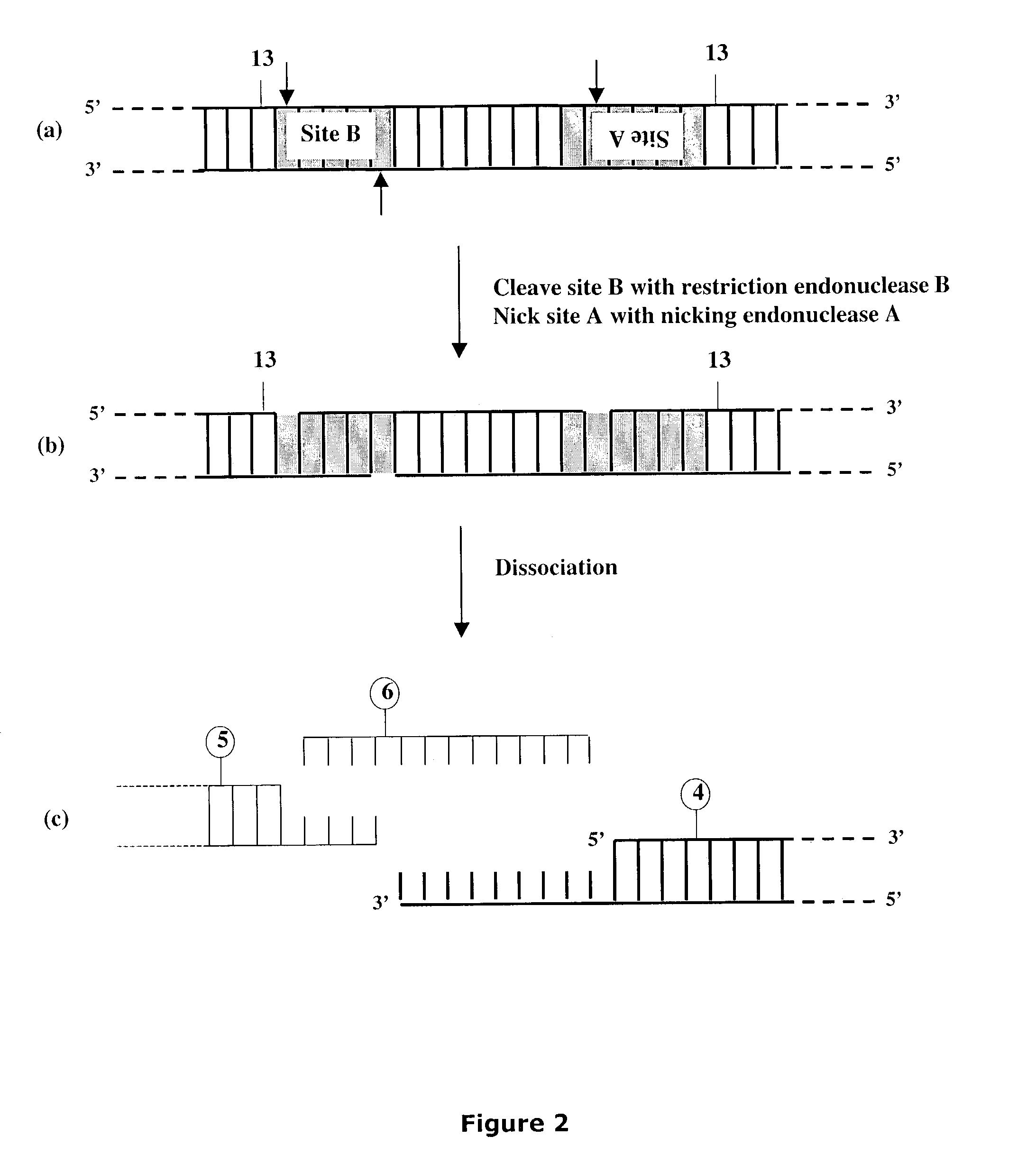
Methods and compositions for DNA manipulation
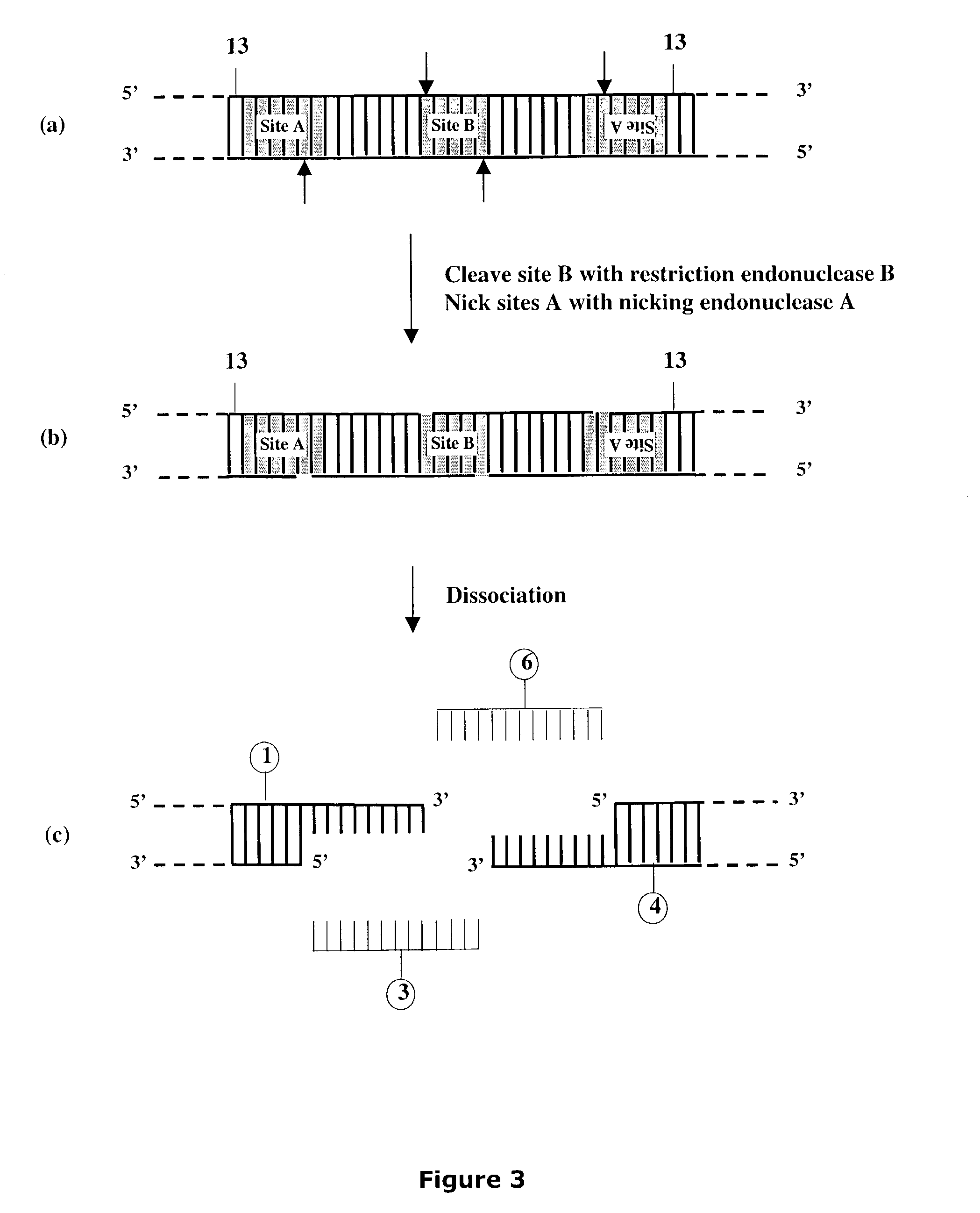
Methods and compositions are provided for generating a single-stranded extension on a polynucleotide molecule, the single-stranded extension having a desired length and sequence composition. Methods for forming single-stranded extensions include: the use of a cassette containing at least one nicking site and at least one restriction site at a predetermined distance from each other and in a predetermined orientation; or primer-dependent amplification which introduces into a polynucleotide molecule, a modified nucleotide which is excised to create a nick using a nicking agent. The methods and compositions provided can be used to manipulate a DNA sequence including introducing site specific mutations into a polynucleotide molecule and for cloning any polynucleotide molecule or set of joined polynucleotide molecules in a recipient molecule such as a vector of choice.
Owner:NEW ENGLAND BIOLABS
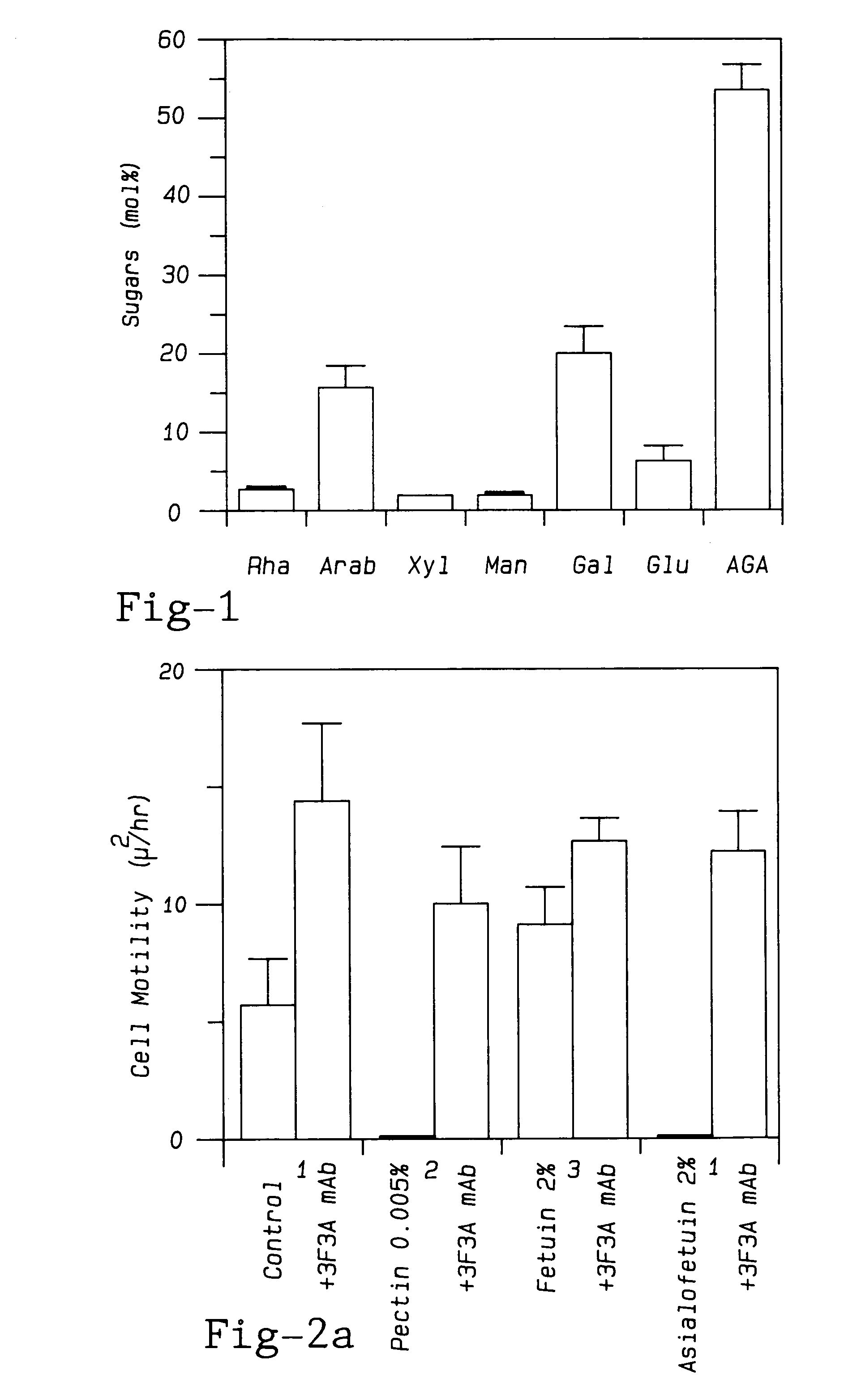
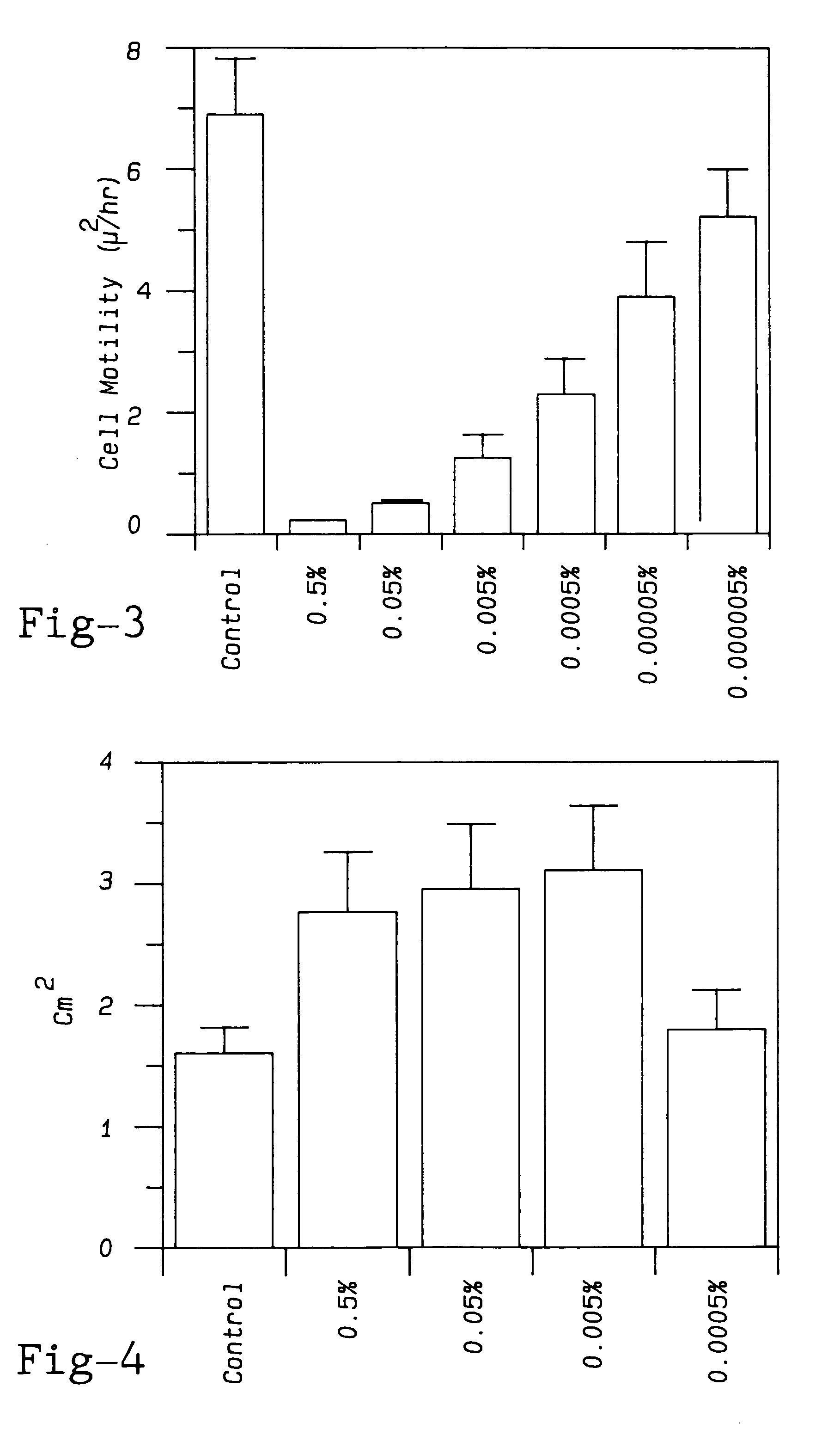
Modified pectin
ActiveUS7491708B1Low degree of branchingInhibit tumor cell migrationOrganic active ingredientsBiocideTumor Cell MigrationSugar
Owner:OXYGEN THERAPY INC
Methods of Clustering Gene and Protein Sequences
InactiveUS20090327170A1Less computationally intensiveReduce computing loadOrganic active ingredientsPeptide/protein ingredientsAntigenNetwork topology
The invention relates to methods for clustering gene and protein sequences. In particular, it involves generation of networks of sequences where the interconnections are based upon a measure of similarity. The invention also provides methods of optimizing and improving the networks by re-wiring of the network based upon overlap of the nearest neighbors of given pairs of nodes. The invention further provides methods of identifying clusters of sequences within the networks and the optimized networks based upon the topology of the network. The clusters identified represent groups of sequences that are related by function and / or evolution. The invention has particular applicability in annotation of sequences in databases and identification of functional homologs which can be very useful for novel therapeutic and diagnostic targets based upon such targets belonging to a cluster or family that contains a known sequence such as a diagnostic sequence, antigen or other therapeutic target.
Owner:DONATI CLAUDIO +2
Detection of HPV
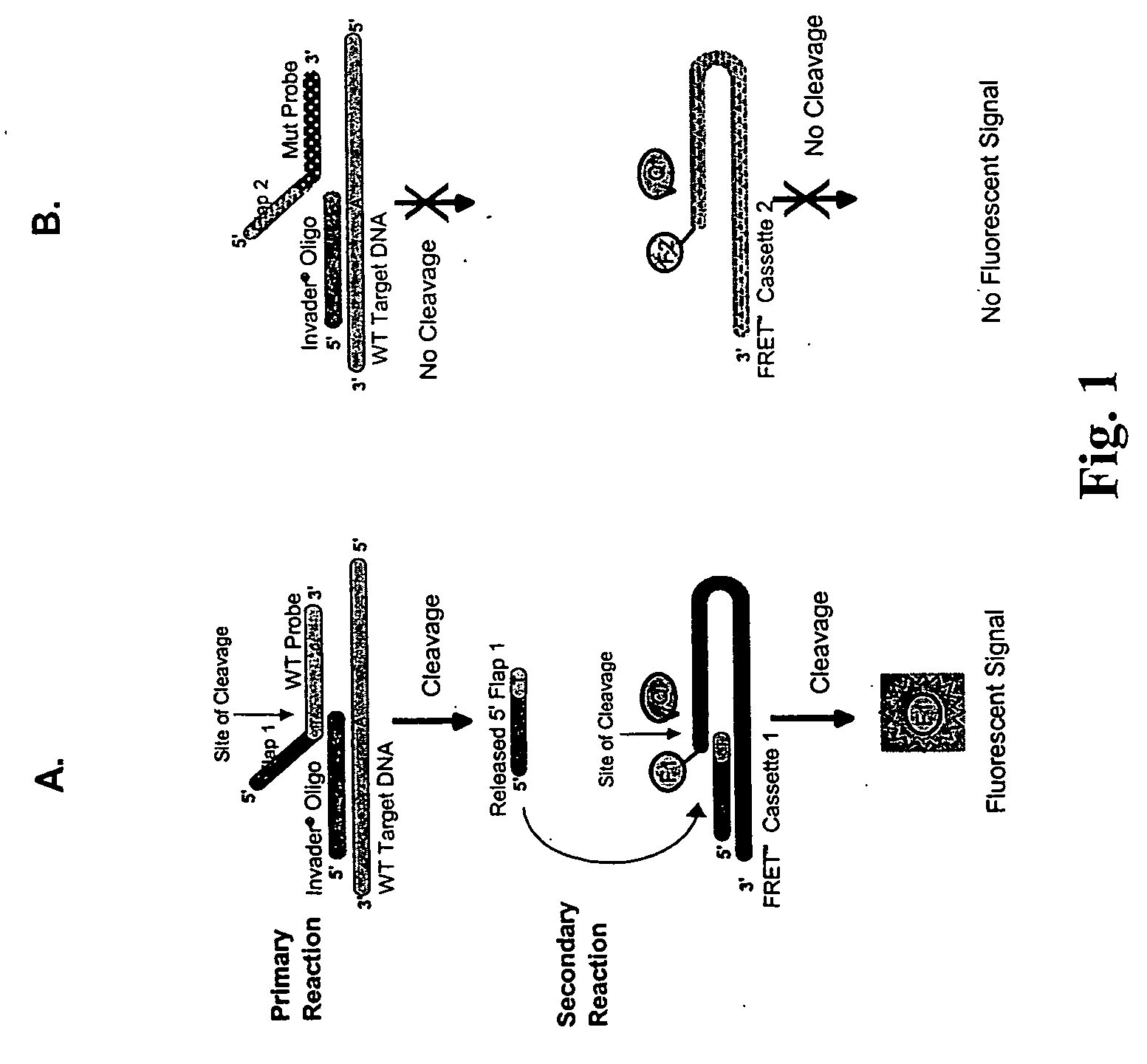
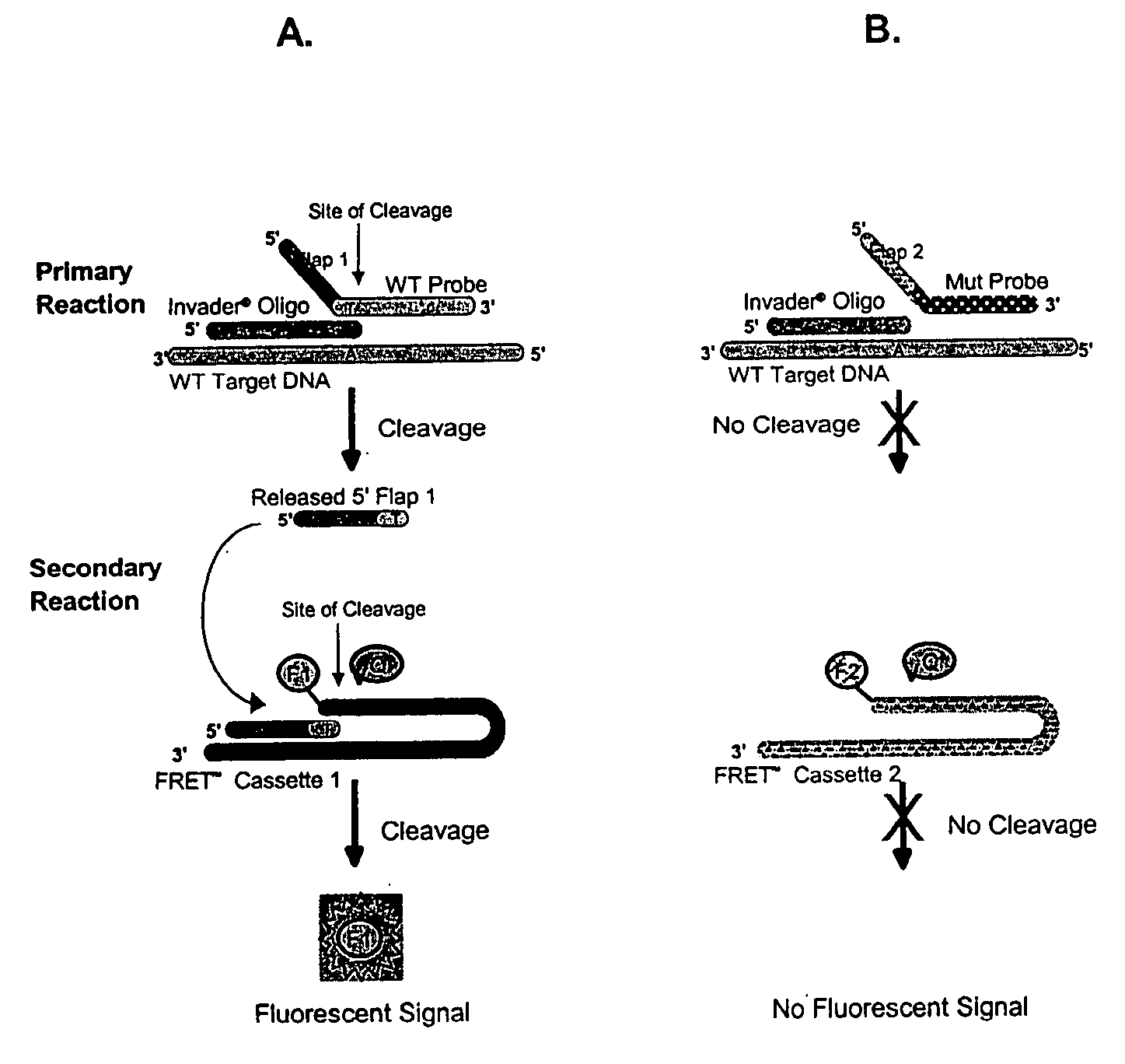
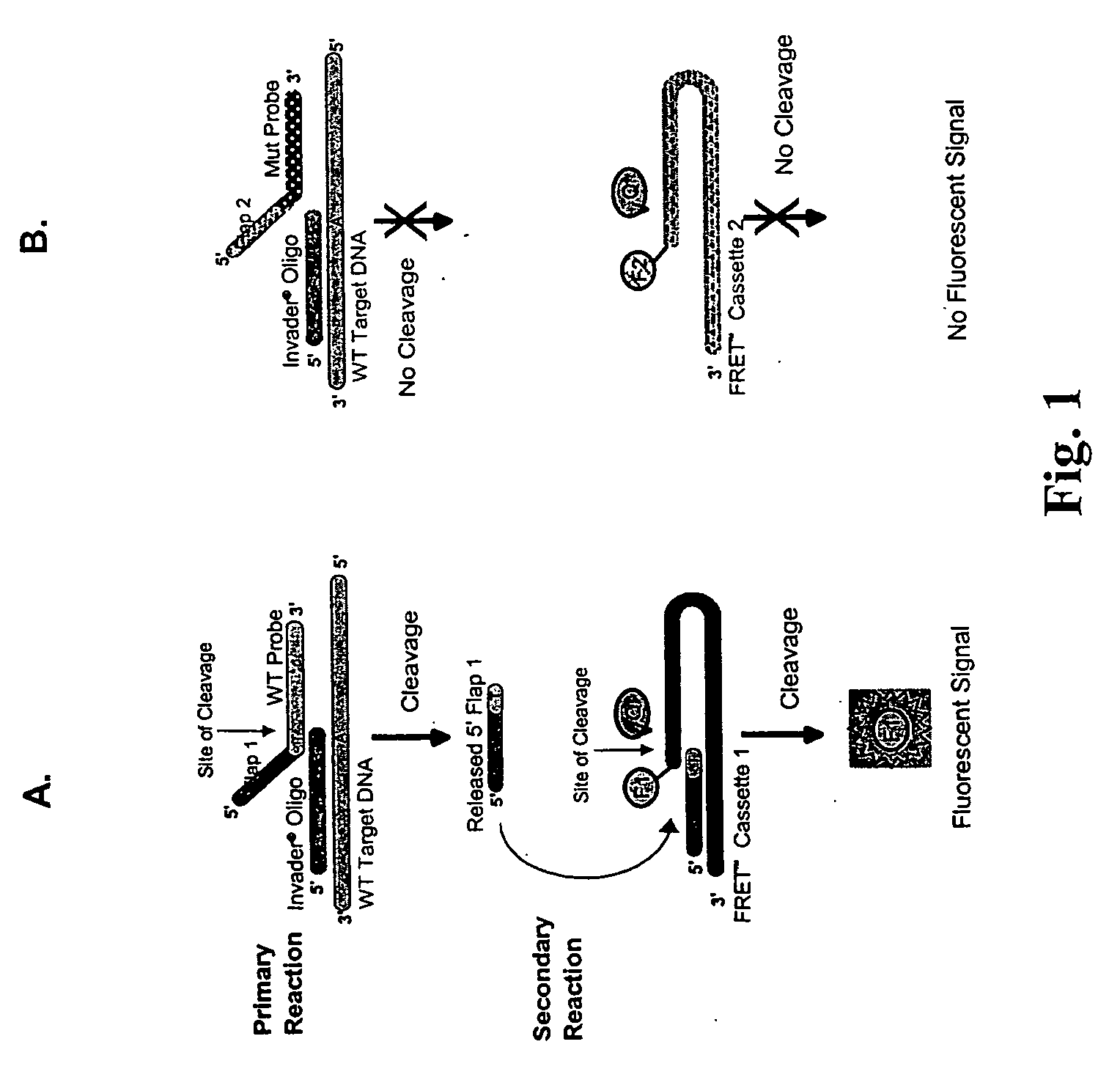
The present invention provides compositions and methods for the detection and characterization of HPV sequences. More particularly, the present invention provides compositions, methods and kits for using invasive cleavage structure assays (e.g. the INVADER assay) to screen nucleic acid samples, e.g., from patients, for the presence of any one of a collection of HPV sequences. The present invention also provides compositions, methods and kits for screening sets of HPV sequences in a single reaction container.
Owner:GEN PROBE INC
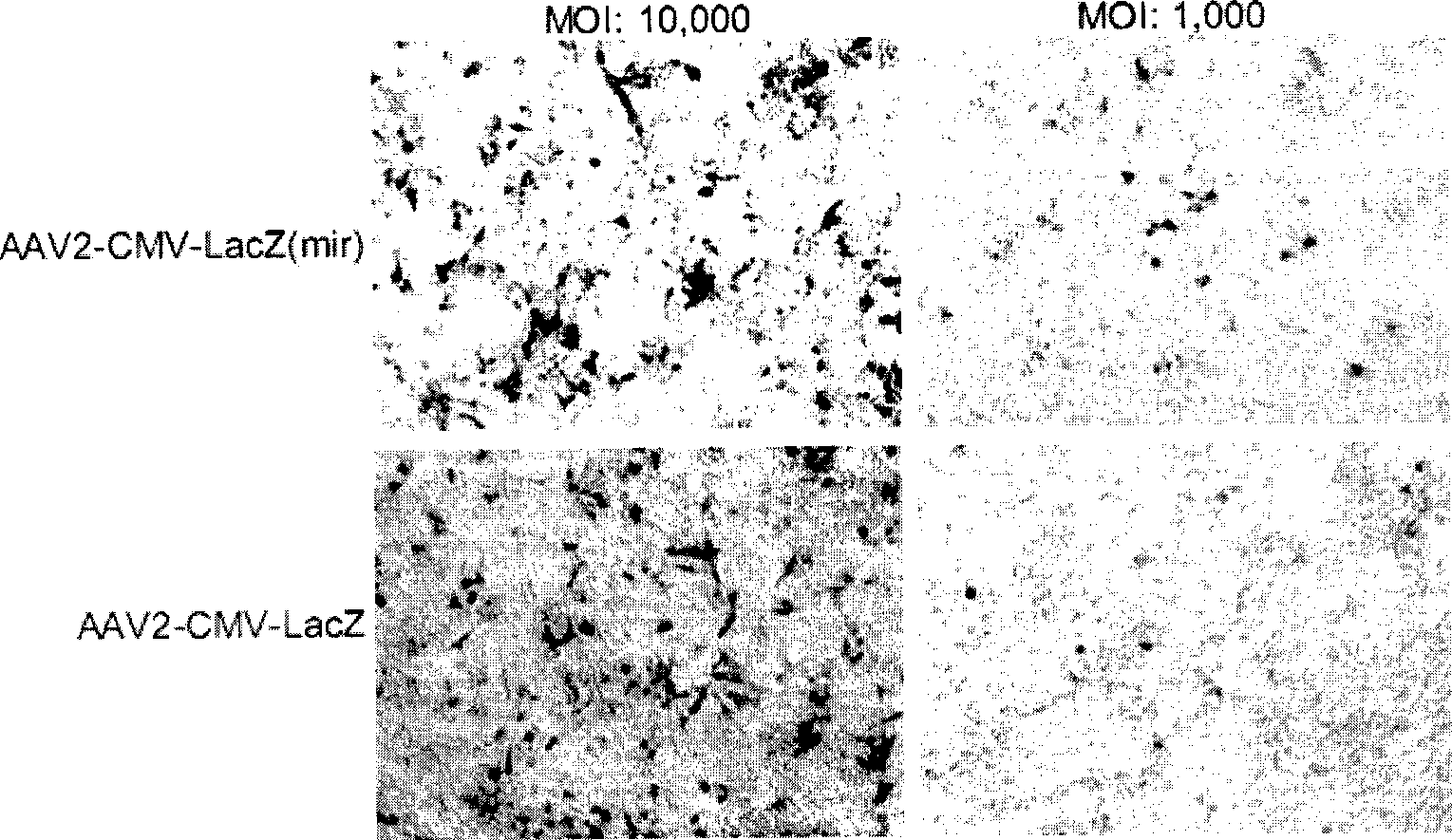
New type cell special gene HAAVmir containing microRNA combined sequence for gene treating
InactiveCN101532024ADoes not affect appearanceDoes not affect productivityGenetic material ingredientsMicroorganism based processesMicroRNACell strain
Owner:许瑞安 +2
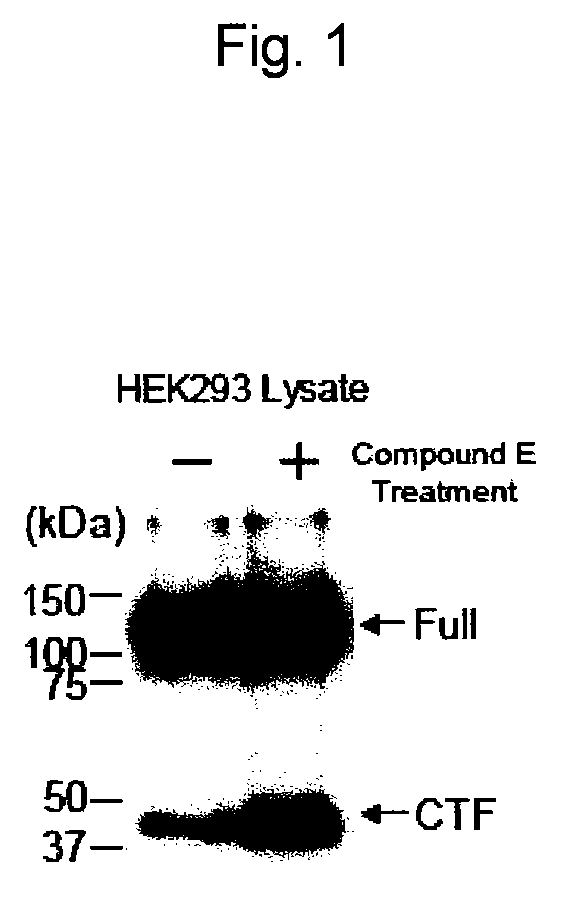
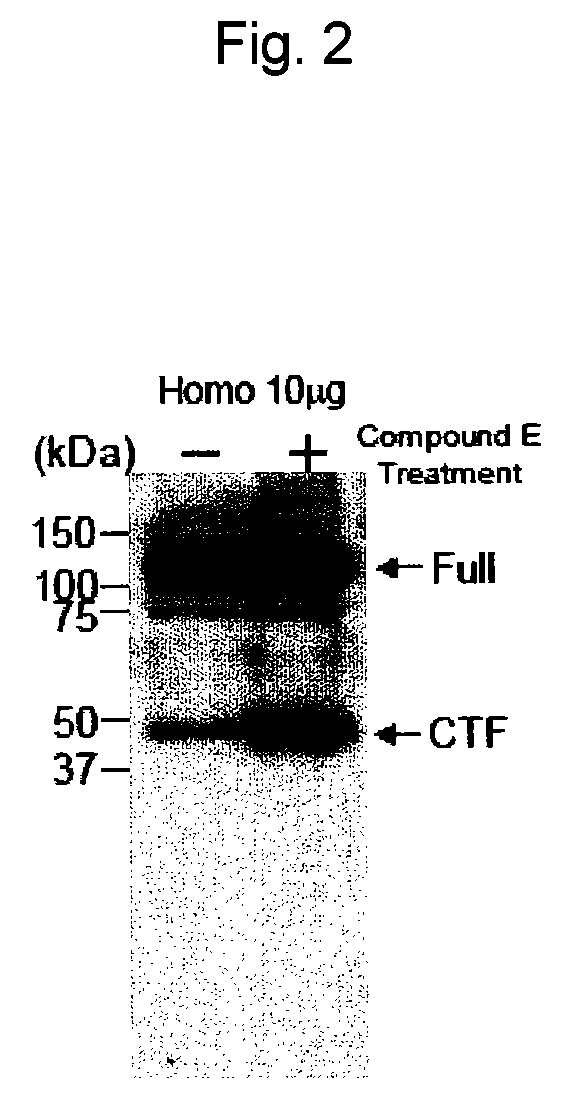
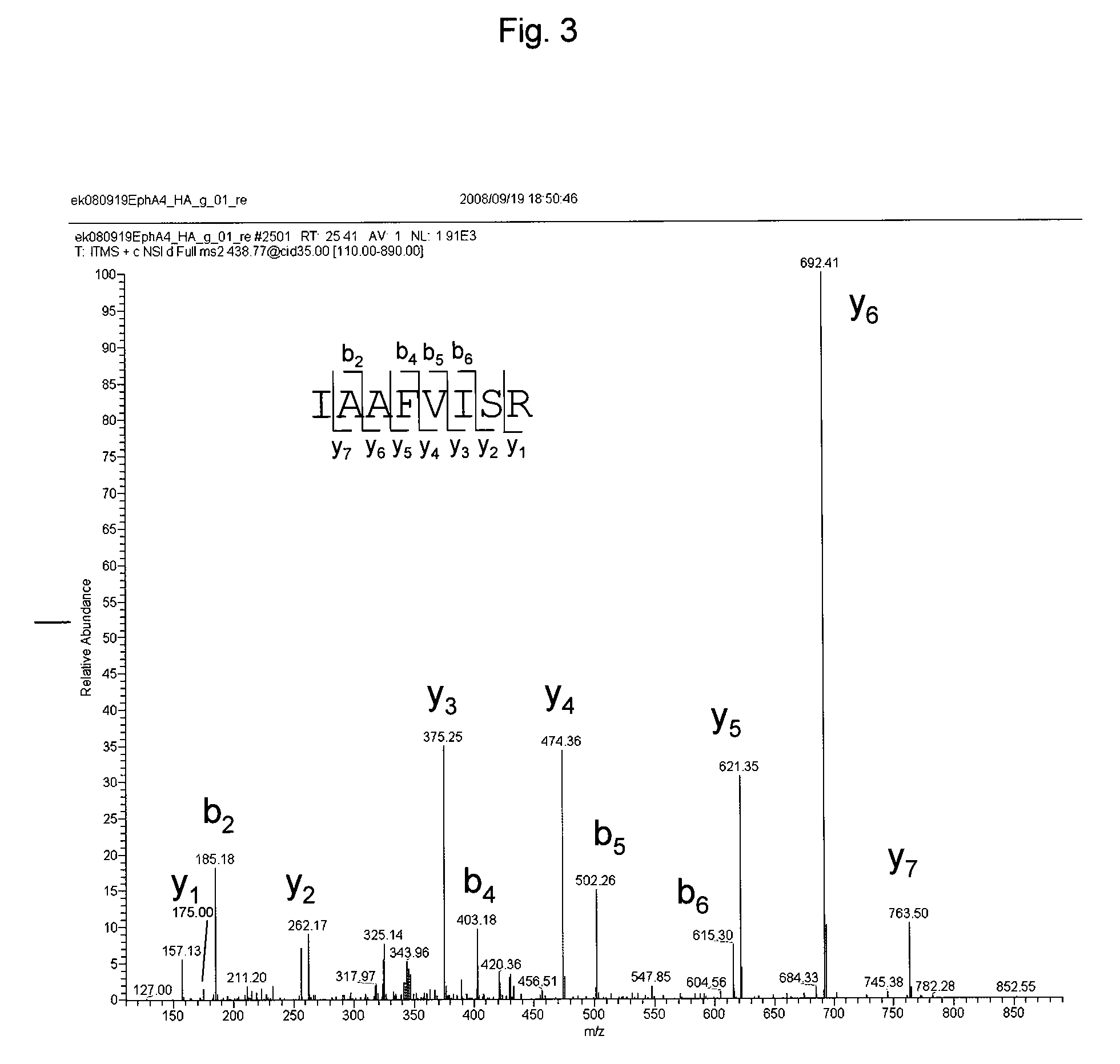
EphA4 POLYPEPTIDE HAVING A NOVEL ACTIVITY AND USE THEREOF
The present invention provides a polynucleotide which consists of a partial sequence of a polynucleotide encoding the amino acid sequence as shown in SEQ ID NO: 2, wherein the former polynucleotide encodes a polypeptide consisting of a partial amino acid sequence of SEQ ID NO: 2 whose N-terminus starts between positions 564 and 621 of SEQ ID NO: 2.
Owner:EISIA R&D MANAGEMENT CO LTD
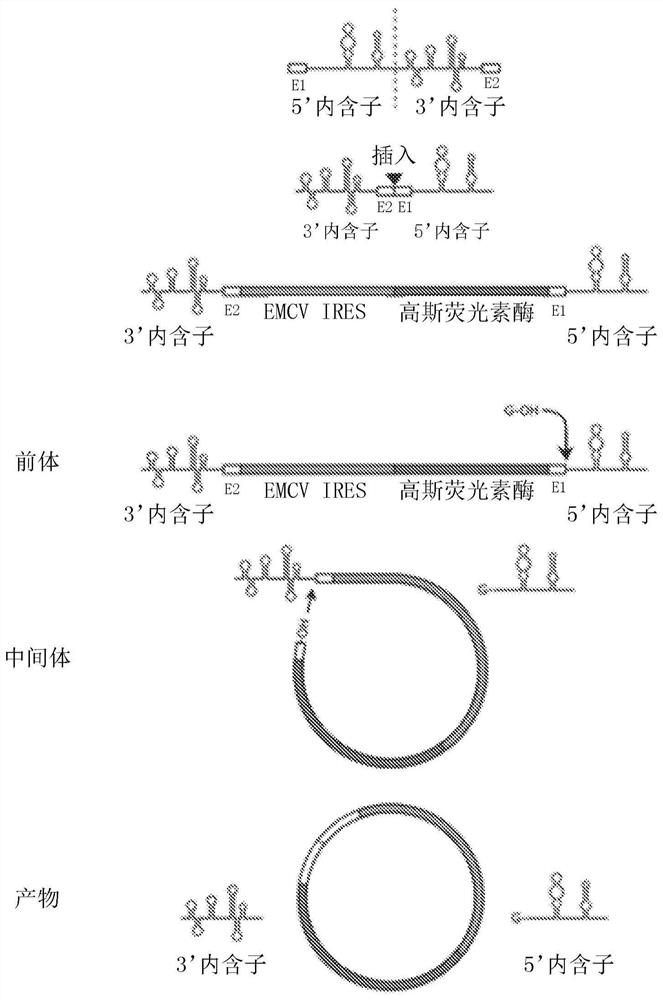
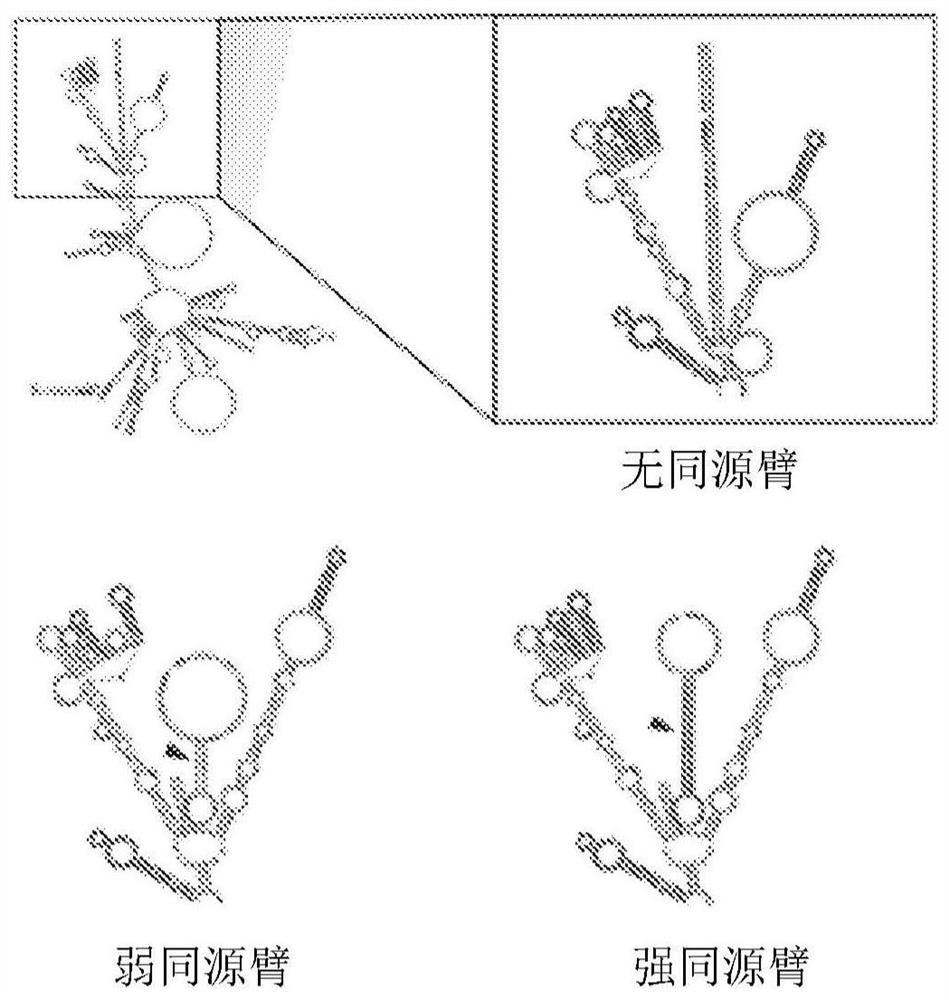
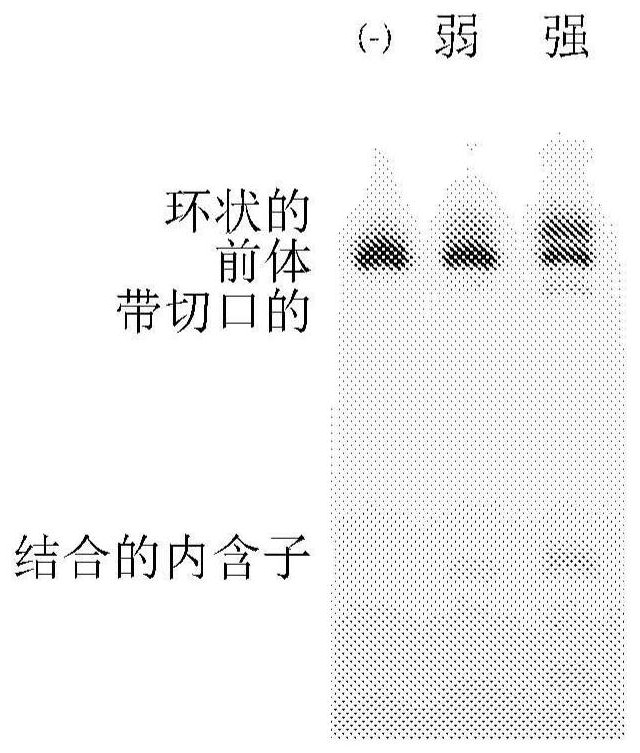
Circular RNA for translation in eukaryotic cells
Owner:MASSACHUSETTS INST OF TECH
Detection Of HPV
The present invention provides compositions and methods for the detection and characterization of HPV sequences. More particularly, the present invention provides compositions, methods and kits for using invasive cleavage structure assays (e.g. the INVADER assay) to screen nucleic acid samples, e.g., from patients, for the presence of any one of a collection of HPV sequences. The present invention also provides compositions, methods and kits for screening sets of HPV sequences in a single reaction container.
Owner:GEN PROBE INC
Polypeptide chip and application thereof in virus detection
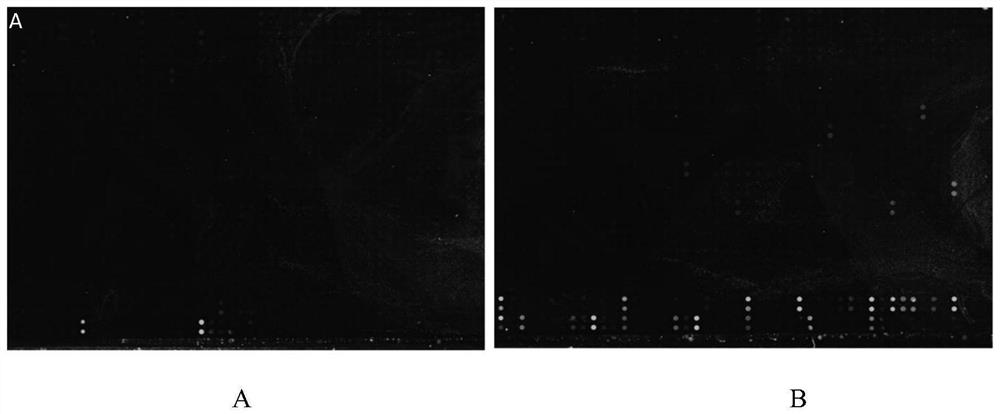
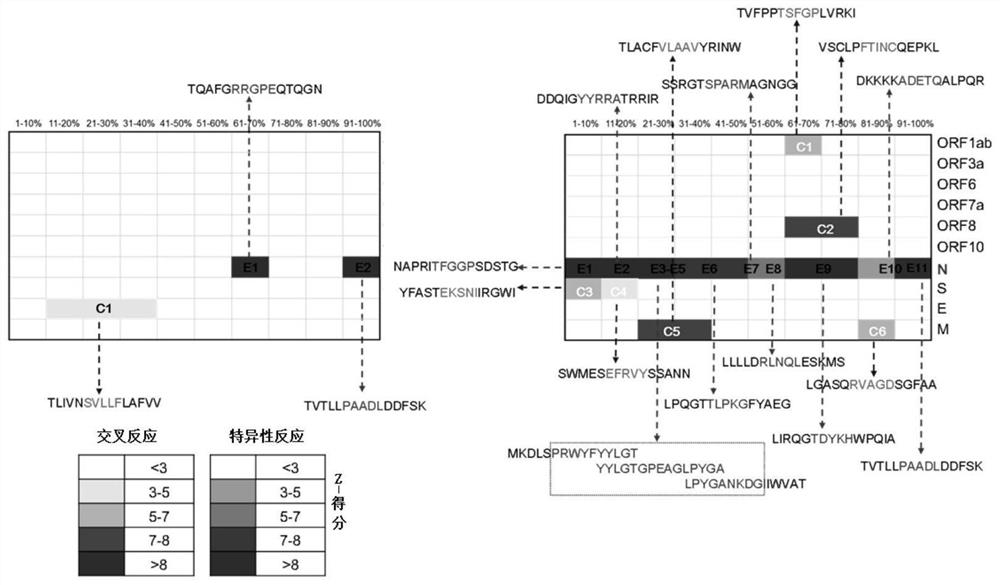
The invention provides a polypeptide chip, which comprises a substrate and n polypeptides distributed on the substrate in an array, each polypeptide sequence has 10-20 amino acids, a group consistingof sequences of first to nth polypeptides covers at least 95% of a viral protein sequence, the adjacent polypeptides have an overlap of 3-8 amino acids, and n is 810-1370. According to the method, proteomics and system biology strategies are adopted, all coded protein sequences of COVID-19 are extracted from NCBI data, an SARS-CoV-2 virus proteome polypeptide chip is designed and prepared, and panoramic scanning of all SARS-CoV-2 virus antibodies in blood of a novel pneumonia virus infected patient is achieved.
Owner:ACADEMY OF MILITARY MEDICAL SCI
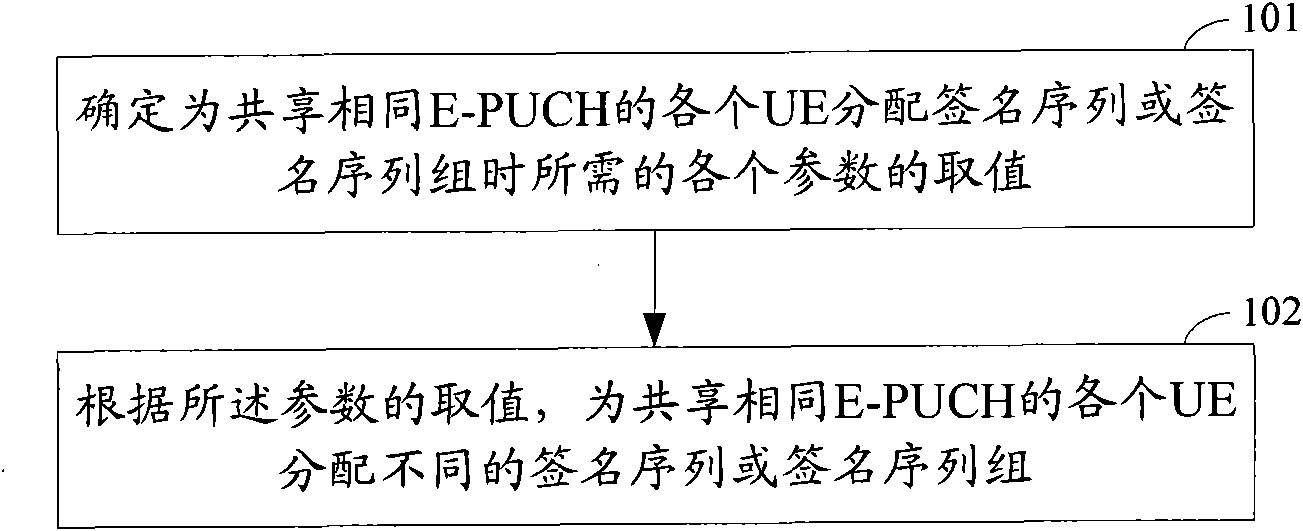
Distribution method of signature sequence or signature sequence group on E-HICH
The invention discloses a distribution method of a signature sequence or a signature sequence group on an E-HICH, comprising the following steps of determining a value of each parameter needed when distributing the signature sequence or the signature sequence group to each user equipment UE which shares a same enhanced physical uplink channel (E-PUCH); and distributing corresponding signature sequence or signature sequence group to each UE which shares the same E-PUCH according to the value of each parameter. By the method, different signature sequences or different signature sequence groupsare distributed to each UE sharing the same E-PUCH, and therefore, NODEB can respectively feed back control information of the E-PUCH of each UE to each UE sharing the same E-PUCH by distributing thesignature sequence or the signature sequence group to each UE.
Owner:TD TECH COMM TECH LTD
Combined primer for amplification and typing of human papilloma virogenes and application of combined primer
InactiveCN104561372AAchieve accurate typingFew stepsMicrobiological testing/measurementMicroorganism based processesForward primerTyping
The invention discloses a combined primer for the amplification and typing of human papilloma virogenes. The combined primer is divided into a combined forward primer and a combined reverse primer; the combined forward primer is composed of three parts, namely a joint part P2 sequence, a tag sequence and a forward primer sequence in order; the combined reverse primer is composed of a reverse primer sequence and a joint part P1 sequence in order. The invention also discloses a kit for the amplification and typing of the human papilloma virogenes. The invention also discloses the applications of the combined primer and the kit. The combined primer has real flux, and is low in cost and capable of accurately typing HPV. The type of HPV can be accurately determined by using a plurality of primers (6 forward primers and 5 reverse primers) for amplification and comparing the sequence information of the amplification product with an HPV database; as a result, a basis is provided for clinical diagnosis and treatment scheme selection.
Owner:南京普东兴生物科技有限公司
Polynucleotide for identifying male and female ginkgo strains and application thereof
ActiveCN110195068AThe result is stable and accurateGood repeatabilityMicrobiological testing/measurementPlant peptidesPhysiologyNucleotide
The invention provides a polynucleotide for identifying male and female ginkgo strains. The polynucleotide comprises or consists of: 1) a nucleotide sequence shown as SEQ ID NO: 5; or 2) a complement,degenerate or homologous sequence of the sequence shown as SEQ ID NO: 5; or 3) a polynucleotide which hybridizes to the nucleotide sequence shown in SEQ ID NO: 5 under stringent conditions or a complement thereof; 4) a coding sequence of any of the sequences 1) to 3); or 5) a nucleotide sequence identical to a nucleotide of 10 bp or more in any of the sequences of sequences 1) to 4). The polynucleotide provided by the present invention is unique among male ginkgo plants, uses the same to identify male and female plants of ginkgo, has the advantages of stable and accurate results, good reproducibility and rapid detection, and provides a basis for the identification of male and female ginkgo by molecular markers.
Owner:AGRI GENOMICS INST CHINESE ACADEMY OF AGRI SCI +1
Primer set and method for identifying variety authentication and seed purity of tomatoes
ActiveCN106350584AImprove accuracyThe result is stableMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceAuthentication
The invention discloses a primer set and method for identifying variety authentication and seed purity of tomatoes. Sequences of 48 pairs of primers of the primer set for identifying variety authentication of tomatoes and purity of seeds is successively as shown in sequences 1-96 in a sequence table, a sequence 2n and a sequence 2n-1 form a primer pair, and n is a natural number in 1-24. The method for identifying variety authentication and seed purity of tomatoes comprises the following steps: carrying out PCR amplification on DNA of tomatoes to be detected by using the primer set; and determining variety authentication and seed purity of the tomatoes to be detected according to difference of PCR products of the tomatoes to be detected. Experiments prove that the primer set not only can be used for identifying the variety authentication of the tomatoes, but also can be used for identifying the seed purity of the tomatoes, and has the advantages that the accuracy is high, results are stable, the primer set is not affected by environment conditions and development stages, statistics is simple and convenient and the like.
Owner:北京通州国际种业科技有限公司 +1
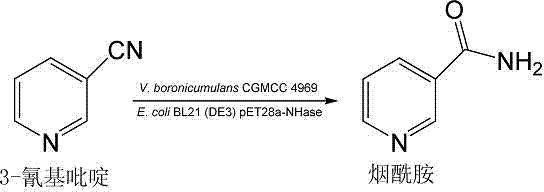
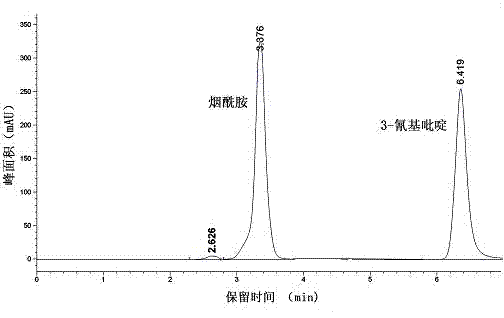
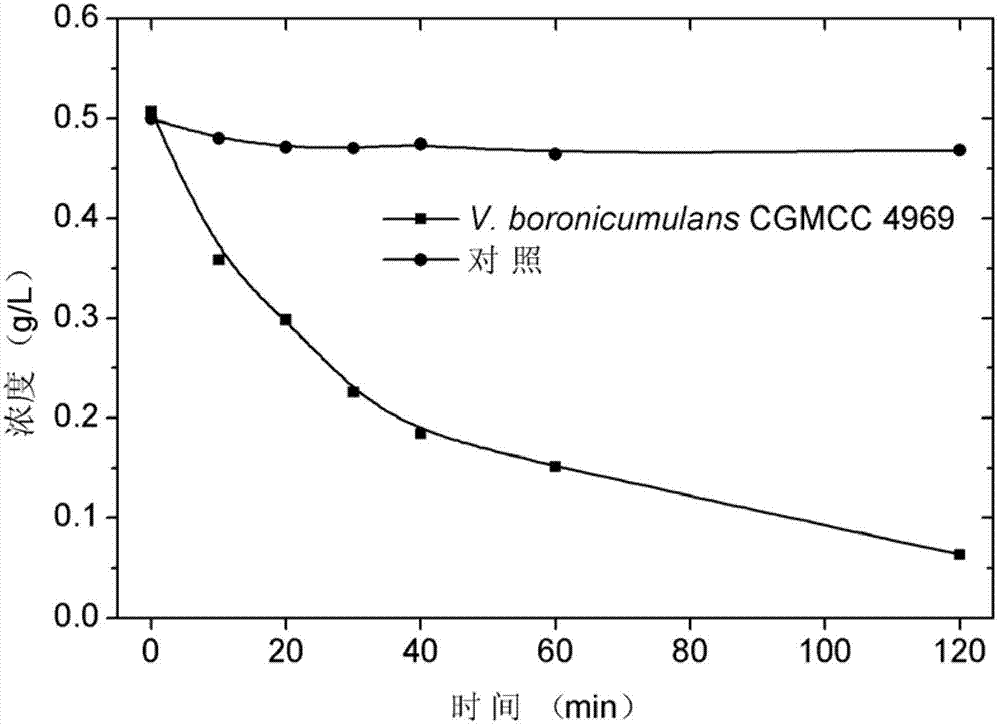
Vaporophage bacterium cgmcc4969 and its application in the biotransformation of 3-cyanopyridine to nicotinamide
The invention discloses the use of variovoraxboronicumulans CGMCC 4969 in bioconversion of 3-cyanopyridine for forming nicotinamide. The nitrile hydratase gene cluster produced by the strain consists of a DNA sequence represented by SEQ ID No.1 in a sequence table; the DNA consisting of the sequence represented by SEQ ID No.1 is recombined onto a pET28a plasmid and can be induced to express in EscherichiacoliBL21(DE3) strain; and the Escherichia coli cells containing expressed proteins and cell extracting solution can convert 3-cyanopyridine into nicotinamide.
Owner:NANJING NORMAL UNIVERSITY
Method for solving waveform sequence-matching problems using multidimensional attractor tokens
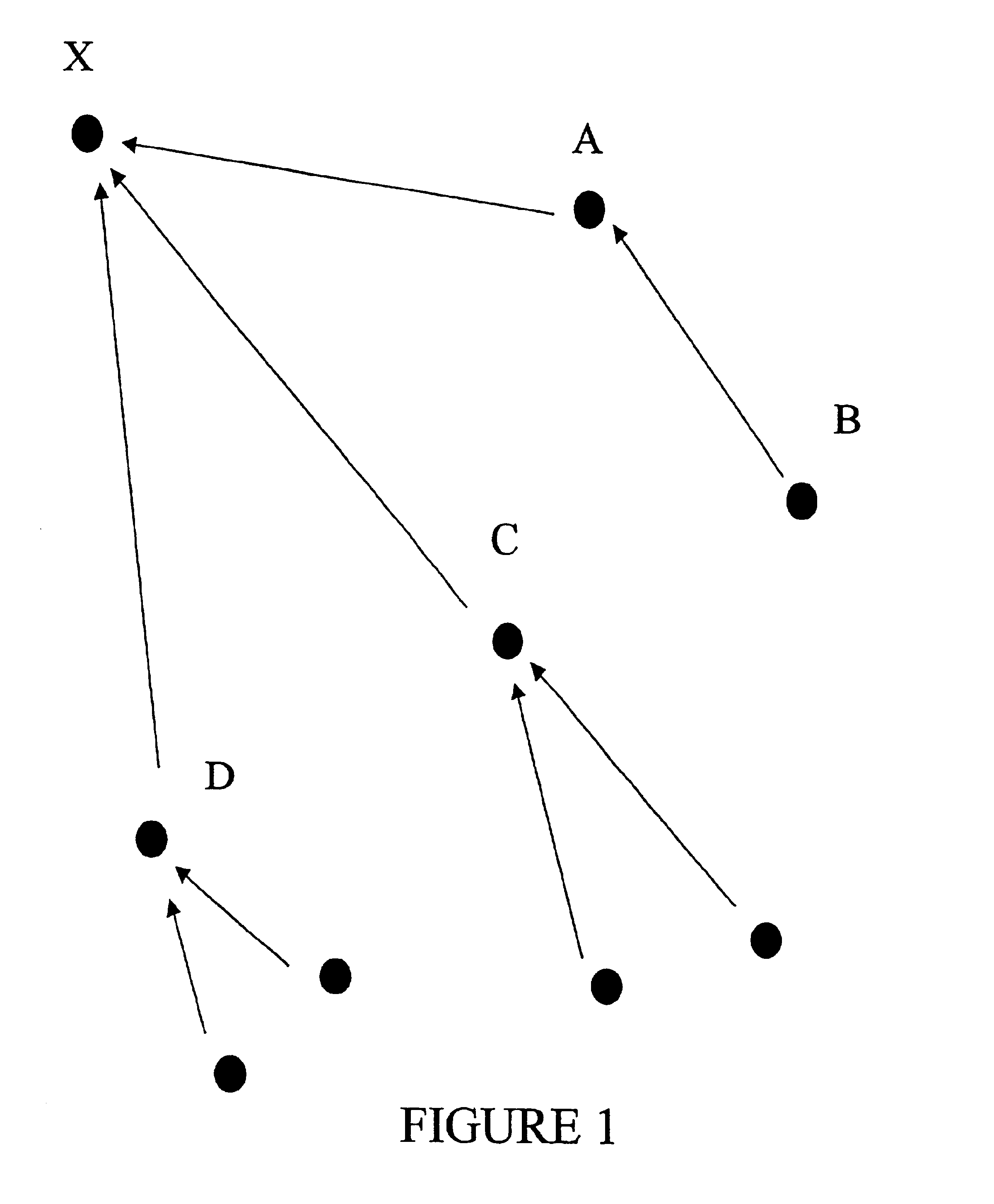
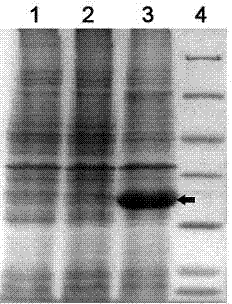
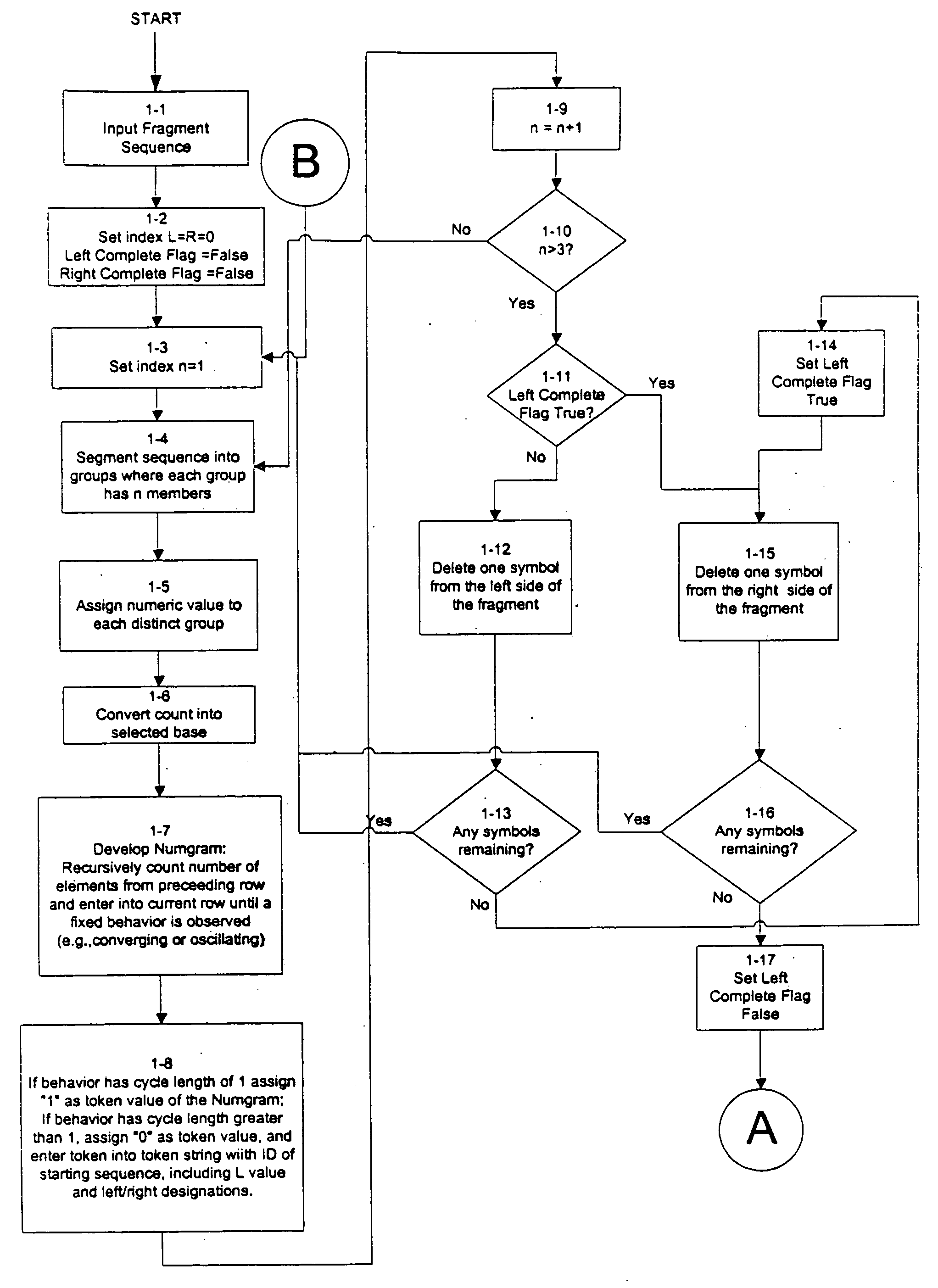
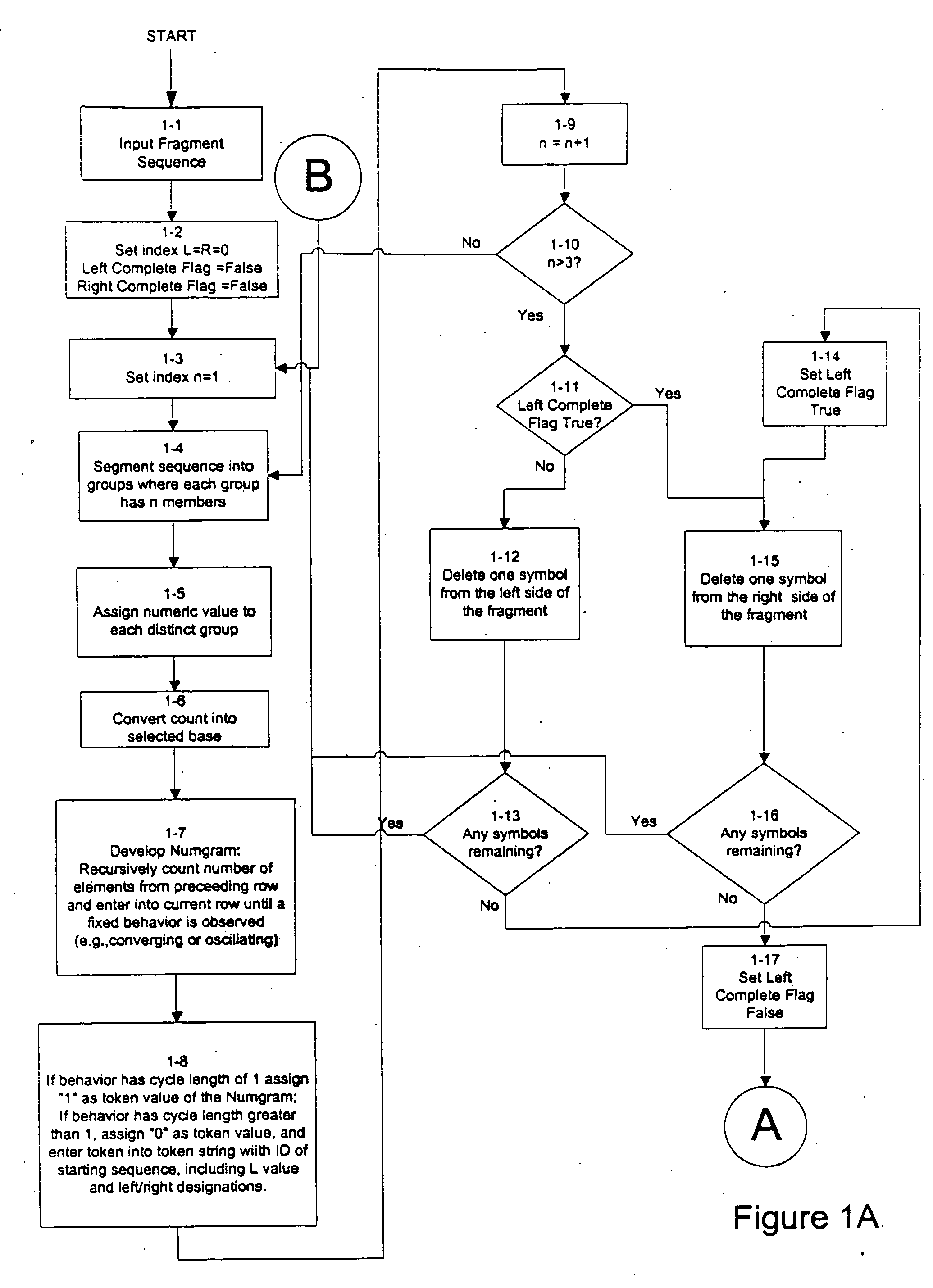
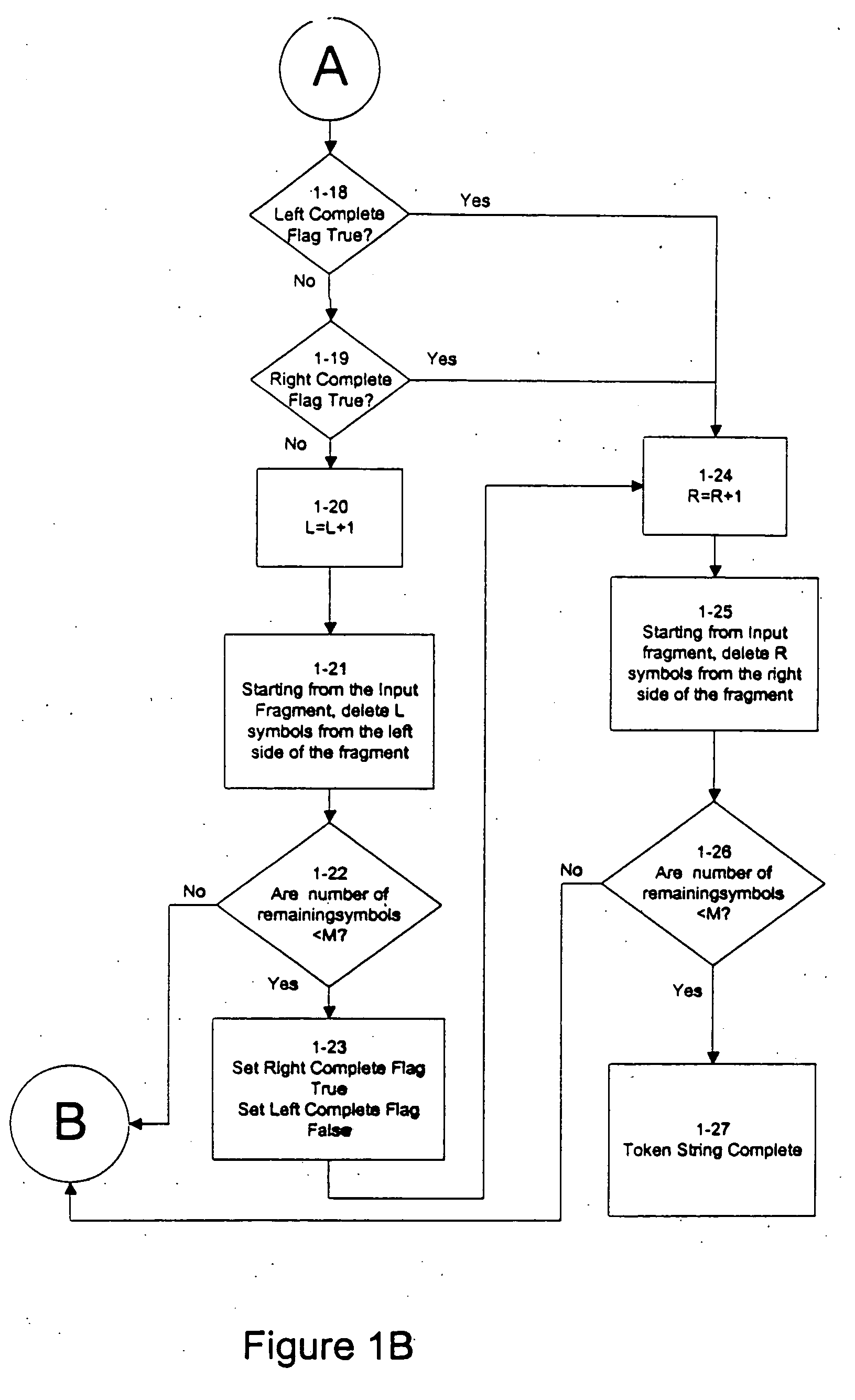
An improved method is provided for solving waveform description, matching and comparison problems using attractor-based processes to extract identity tokens that indicate sequence and subsequence symbol content and order of the waveform or waveform segments. The waveform is described with a suitable alphabet to extract the ontology of the waveform, and syntactical rules are applied to direct pattern extraction using the alphabet. The patterns are extracted in a hierarchical, embedded manner according the global or local maximia and minimia so that the resulting statements are compatible with analysis in catastrophe theory. The attractor processes map the resulting waveform sequence from its original sequence representation space (OSRS) into a hierarchical multidimensional attractor space (HMAS). The HMAS can be configured to represent equivalent symbol distributions within two symbol sequences or perform exact symbol sequence matching. The mapping process results in each sequence being drawn to an attractor in the HMAS. Each attractor within the HMAS forms a unique token for a group of sequences with no overlap between the sequence groups represented by different attractors. The size of the sequence groups represented by a given attractor can be reduced from approximately half of all possible sequences to a much smaller subset of possible sequences. The mapping process is repeated for a given sequence so that tokens are created for the whole sequence and a series of subsequences created by repeatedly removing a symbol or group of symbols from the one end of sequence and then repeating the process from the other end. The resulting string of tokens represents the exact identity of the whole sequence and all its subsequences ordered from each end.
Owner:OMNIGON TECH
Test strip RPA primer and detection kit for detecting Ditylenchus destructor
PendingCN111676296AStrong specificityHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationDitylenchus destructorNucleotide
The invention discloses a test strip RPA primer for detecting Ditylenchus destructor. The primer is composed of nucleotide sequences as shown in SEQ ID No:1 and SEQ ID No:2. The invention further discloses a combination of the primer and a probe, and the probe is composed of a sequence as shown in SEQ ID No:3. In addition, the invention discloses a detection kit comprising the primer and probe combination. The combination of the test strip RPA primer and the probe has the advantages of strong detection specificity for the Ditylenchus destructor, high sensitivity, low requirements on equipmentand personnel quality, simplicity in operation, high detection speed and low cost, and is suitable for port quarantine and basic level on-site detection.
Owner:INST OF PLANT PROTECTION HEBEI ACAD OF AGRI & FORESTRY SCI
CRISPR detection primer group for mycoplasma pneumoniae and application of CRISPR detection primer group
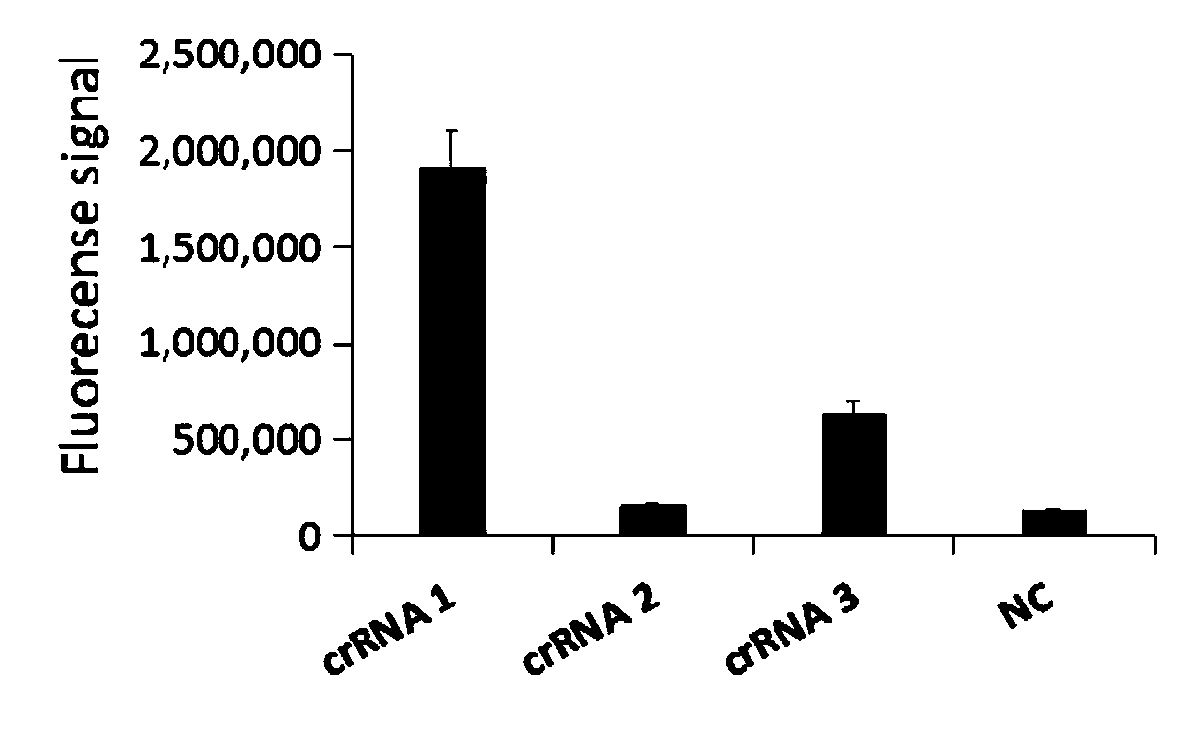
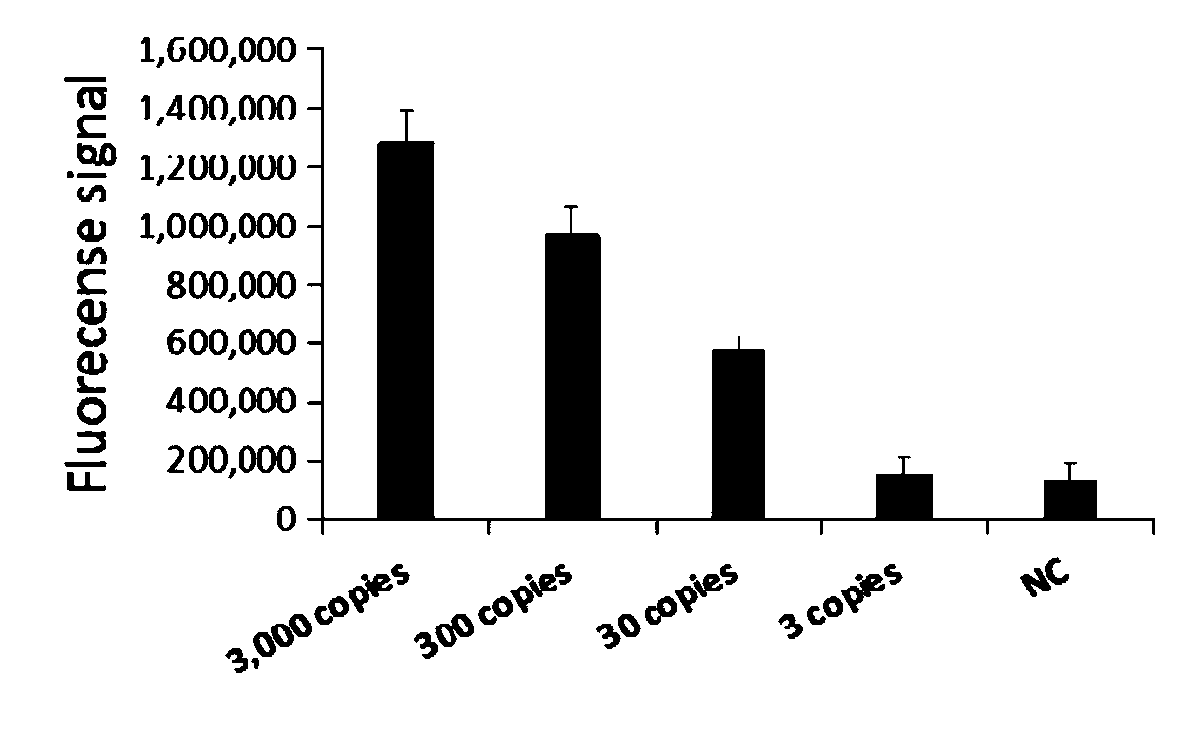
ActiveCN110804669AGet rid of dependenceShorten detection timeMicrobiological testing/measurementMicroorganism based processesMycoplasmaSequenceome
The invention relates to a CRISPR (clustered regularly interspaced short palindromic repeat) detection primer group for mycoplasma pneumoniae and an application of the CRISPR detection primer group, and belongs to the technical field of gene detection of a CRISPR technology. The primer group comprises an amplification primer pair and crRNA (CRISPR-derived ribonucleic acid), wherein the amplification primer pair is used for amplifying a sequence shown as SEQ ID NO. (sequence identifier number) 1 of the mycoplasma pneumoniae; crRNA comprises an anchor sequence and a guide sequence; the anchor sequence specifically identifies a Cas protein; and the guide sequence is matched with a target sequence segment in the sequence of SEQ ID NO. 1. The primer group detects the mycoplasma pneumoniae through the CRISPR technology; the detection time of the mycoplasma pneumoniae is shortened; and the detection can be accomplished within 60min. A specific sequence combination obtained by screening acts as the primer group for detection, the primer group has the advantages of high sensitivity and strong specificity, and a limit of detection of the primer group can reach 30 copies. The primer group isused for the CRISPR detection of the mycoplasma pneumoniae, is independent of a complicated variable temperature amplification instrument such as a qPCR (quantitative polymerase chain reaction) instrument, and a CRISPR-Cas technology has wide application prospects in instant diagnosis of the mycoplasma pneumoniae.
Owner:广州微远医疗器械有限公司 +3
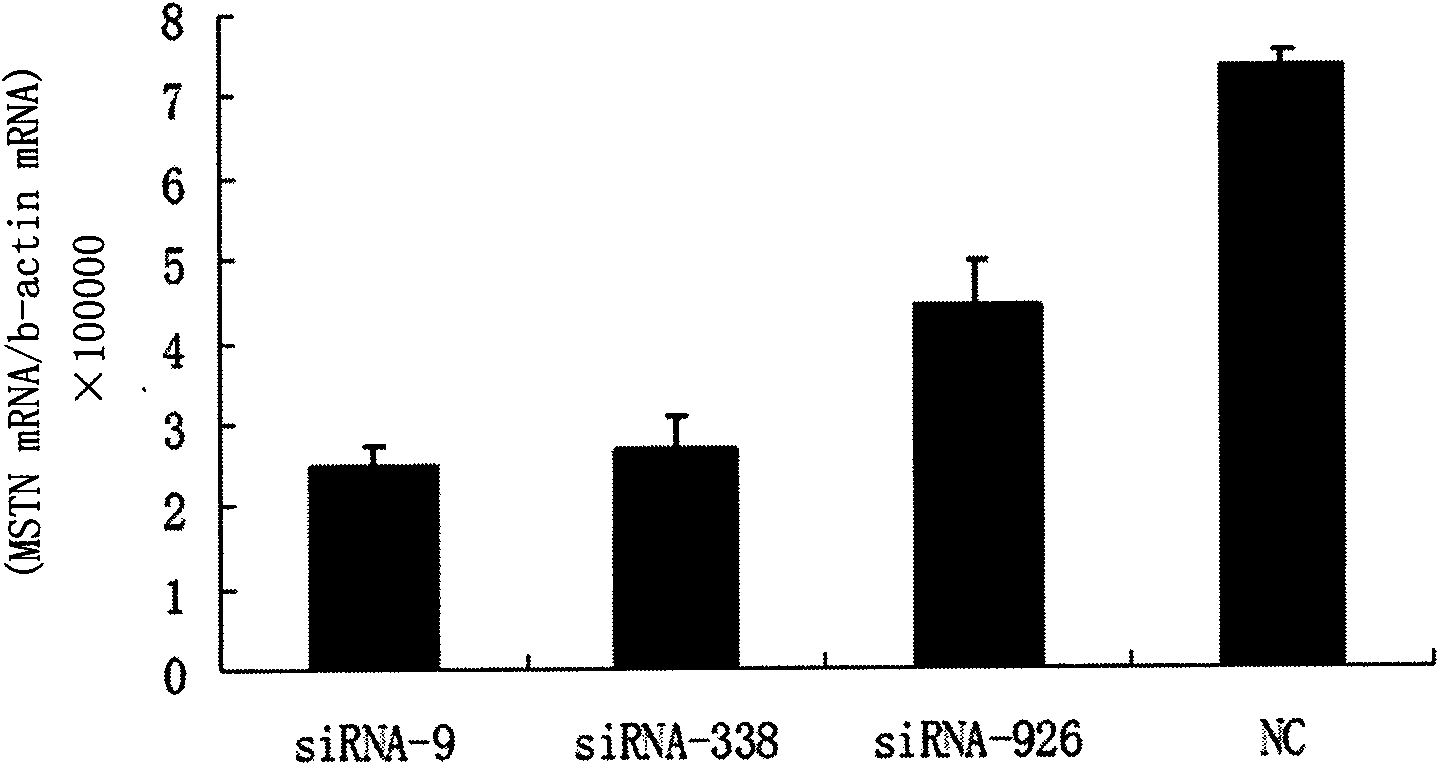
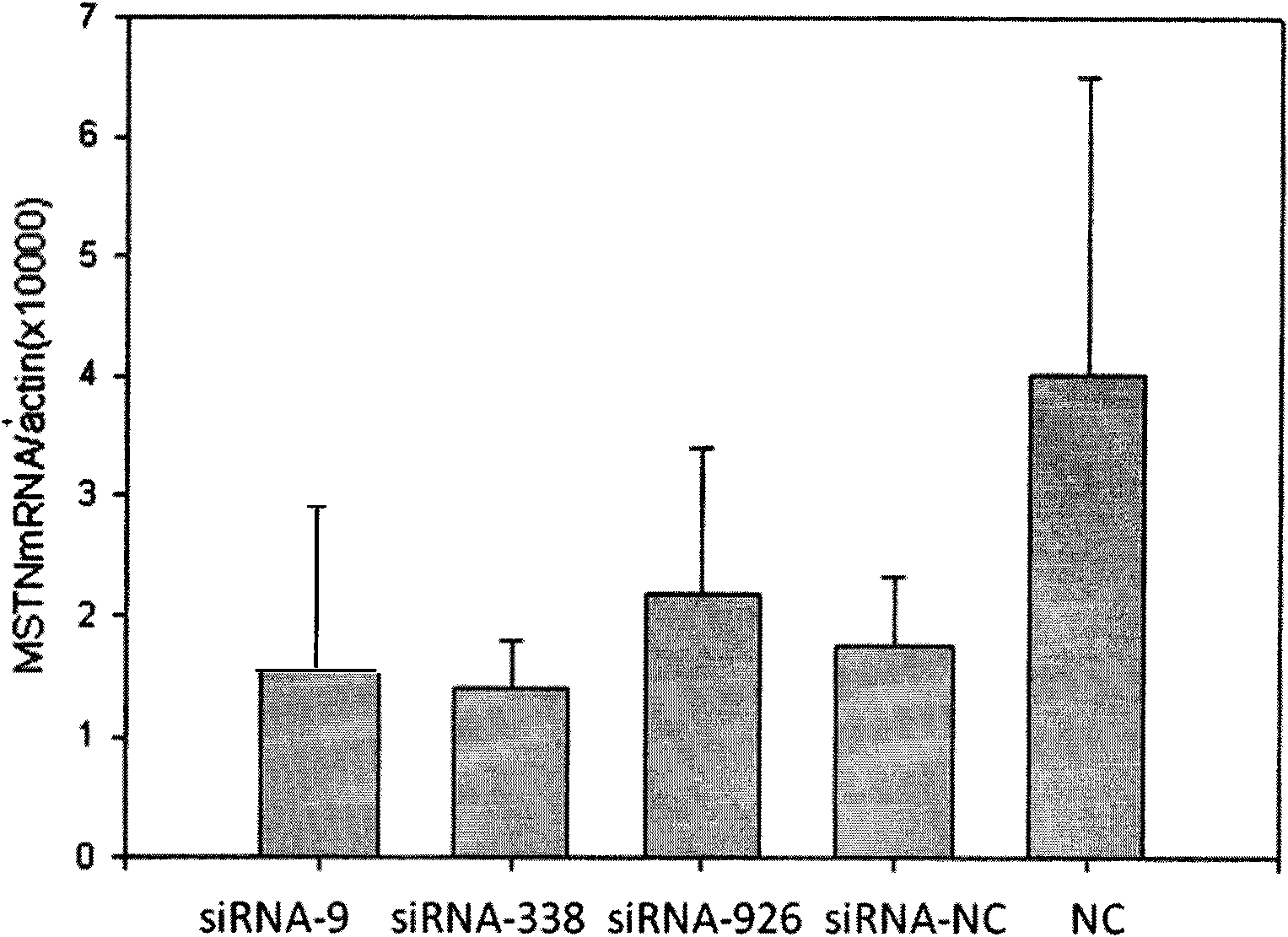
Small interfering RNA (siRNA) inhibiting expression of myostatin (MSTN) gene in chicken and application thereof
InactiveCN102041257ASkeletal muscle enlargementIncrease meat productionMicroencapsulation basedDNA/RNA fragmentationMyostatinBreeding chicken
The invention discloses a small interfering RNA (siRNA) inhibiting expression of a myostatin (MSTN) gene in chicken and application thereof. The invention provides an siRNA which acts on the MSTN gene, wherein the siRNA is a) a double-stranded RNA composed of the sequence 1 and the sequence 2 in a sequence table, b) a double-stranded RNA composed of the sequence 3 and the sequence 4 in the sequence table or c) a double-stranded RNA composed of the sequence 5 and the sequence 6 in the sequence table. The invention obtains the siRNA effectively inhibiting expression of the MSTN gene in chicken,real-time quantitative polymerase chain reaction (PCR) verifies that the siRNA effectively reduces expression of MSTN in molecular biology, and chicken with skeletal muscles obviously increased are successively obtained through microinjection test in embryology. The above results show that the siRNA provided by the invention can be used for breeding chicken, thus improving the meat yields of broilers, especially high quality chicken.
Owner:CHINA AGRI UNIV
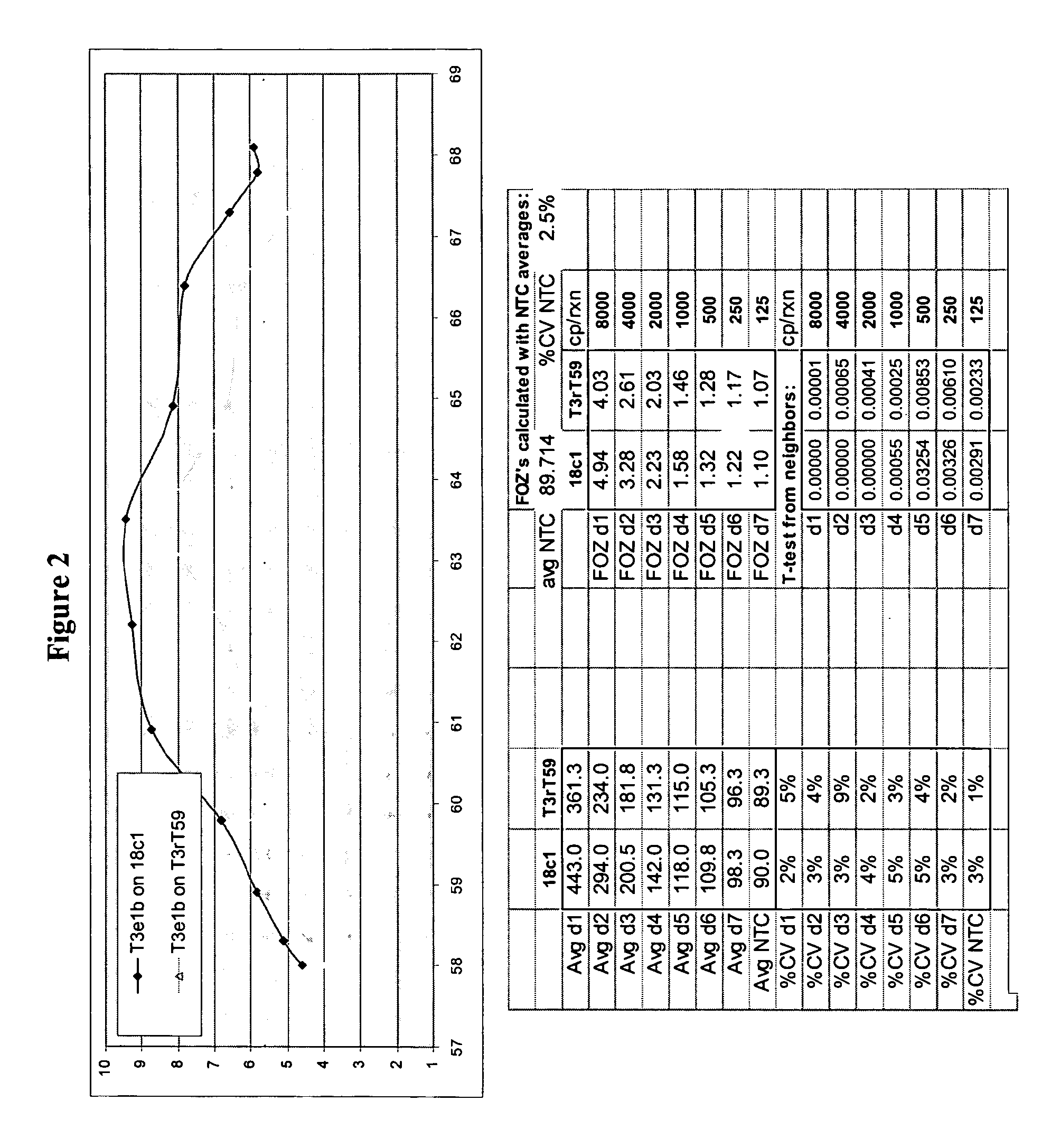
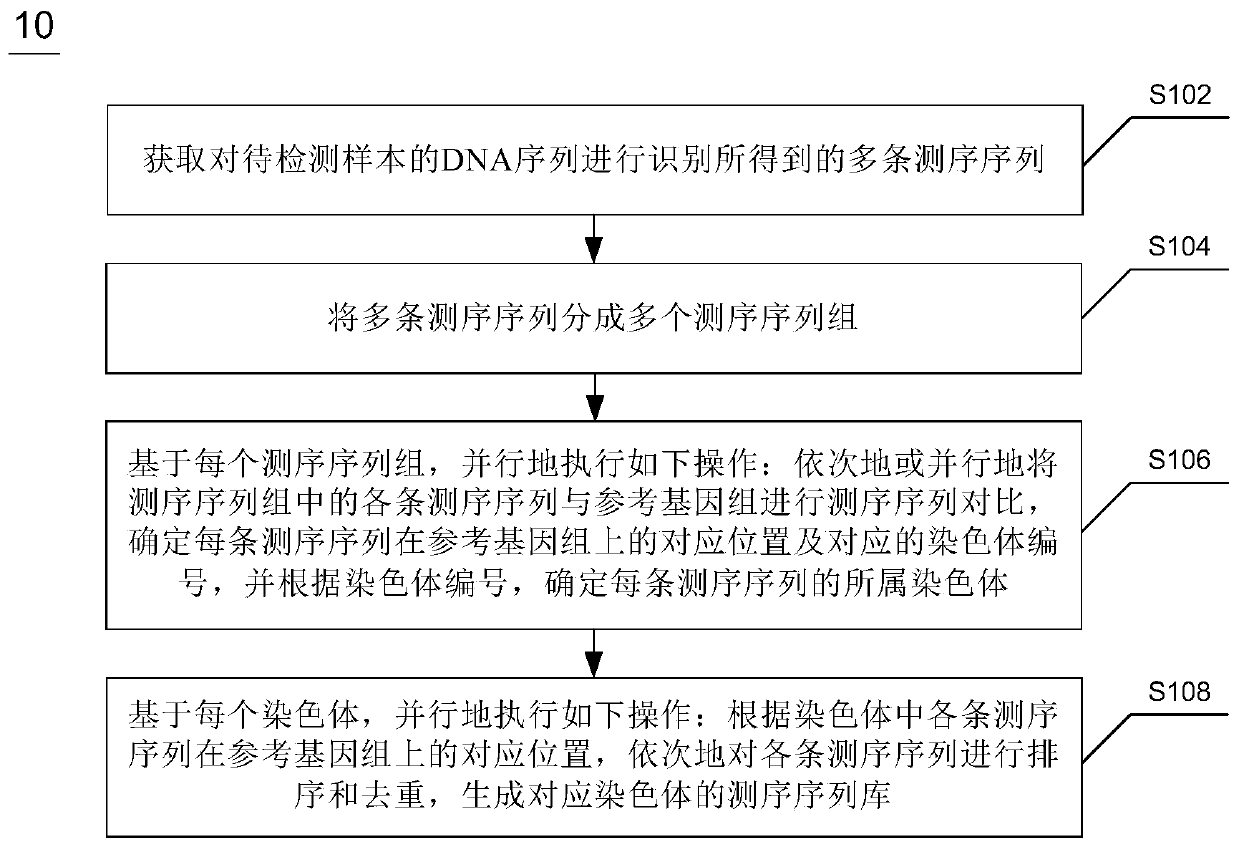
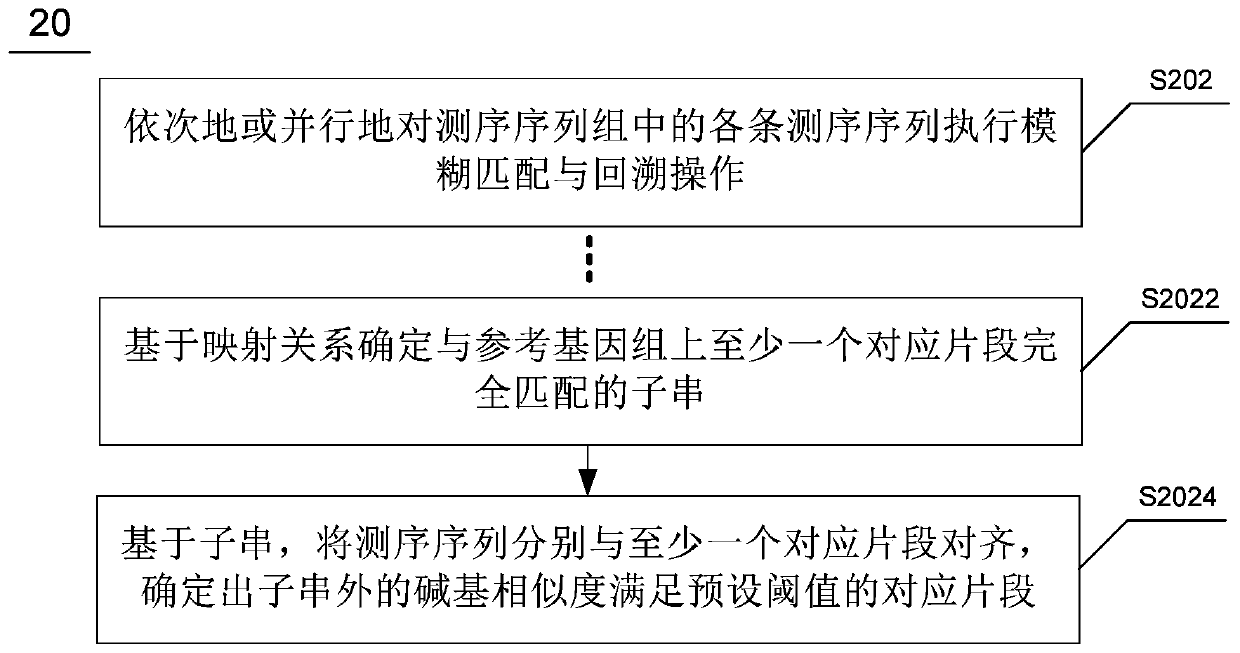
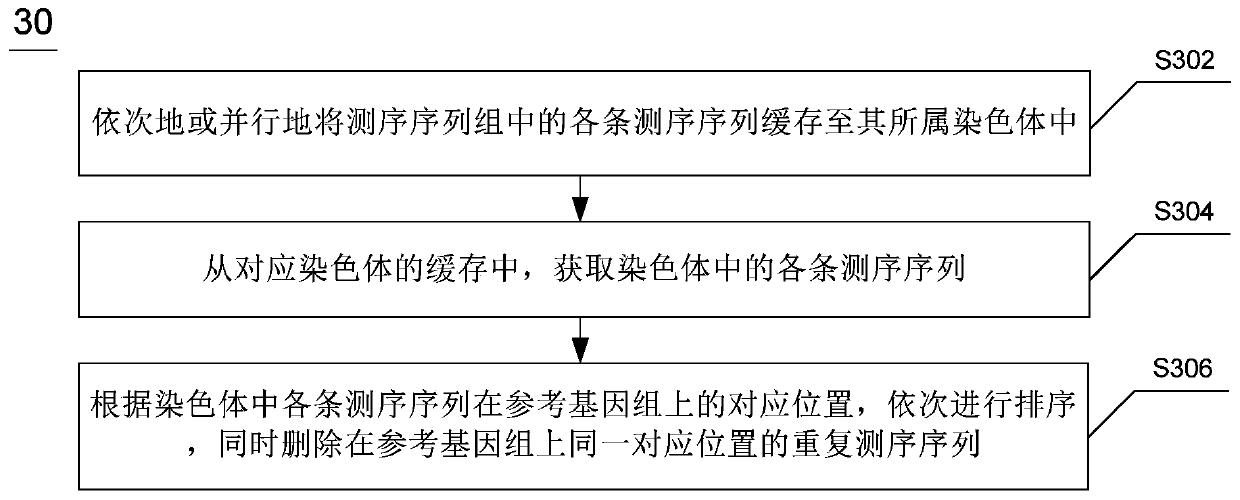
Whole genome re-sequencing analysis and method for whole genome re-sequencing analysis
ActiveCN110797088ATake advantage ofShorten the timeBiostatisticsProteomicsSequence analysisGenome resequencing
The invention discloses whole genome re-sequencing analysis and a method for the whole genome re-sequencing analysis. The method for whole genome re-sequencing analysis comprises the following steps of: acquiring a plurality of sequencing sequences obtained by identifying a DNA sequence of a sample to be detected, dividing the plurality of sequencing sequences into a plurality of sequencing sequence groups, based on each sequencing sequence group, executing the following operations in parallel: sequentially or parallelly comparing each sequencing sequence in the sequencing sequence group witha reference genome, and determining a corresponding position of each sequencing sequence on the reference genome and a corresponding chromosome number, and according to the corresponding position of each sequencing sequence on the reference genome and the corresponding chromosome number, conducting sequencing and duplicate removal on each sequencing sequence, and generating a sequencing sequence library corresponding to each chromosome.
Owner:南京医基云医疗数据研究院有限公司
Method of detecting or quantitating endogenous wheat DNA and method of determining contamination rate of genetically modified wheat in test sample
ActiveUS8173400B2Good precisionSugar derivativesMicrobiological testing/measurementNucleotideTransgene
An object of the present invention is to discover an endogenous wheat sequence satisfying the conditions of: a) it is universally present in varieties of wheat, b) the amount present (detected amount) is not affected by the wheat variety, c) even if other grains are present, only wheat can be detected without cross-reactivity, and d) it is amplified quantitatively by the PCR reaction. A further object of the present invention is to provide a method of accurately detecting and quantitating endogenous wheat DNA in a test sample by the polymerase chain reaction. The present invention provides a method of detecting or quantitating endogenous wheat DNA in a test sample by the polymerase chain reaction, the method comprising: a step of using a nucleic acid molecule in the test sample or a nucleic acid molecule extracted from the test sample as a template to amplify the nucleic acid molecule of a region consisting of the nucleotide sequence identified as SEQ ID NO: 2 or a partial sequence thereof with a primer pair capable of amplifying that region; and a step of detecting or quantitating the amplified nucleic acid molecule.
Owner:NISSHIN SEIFUN GRP INC
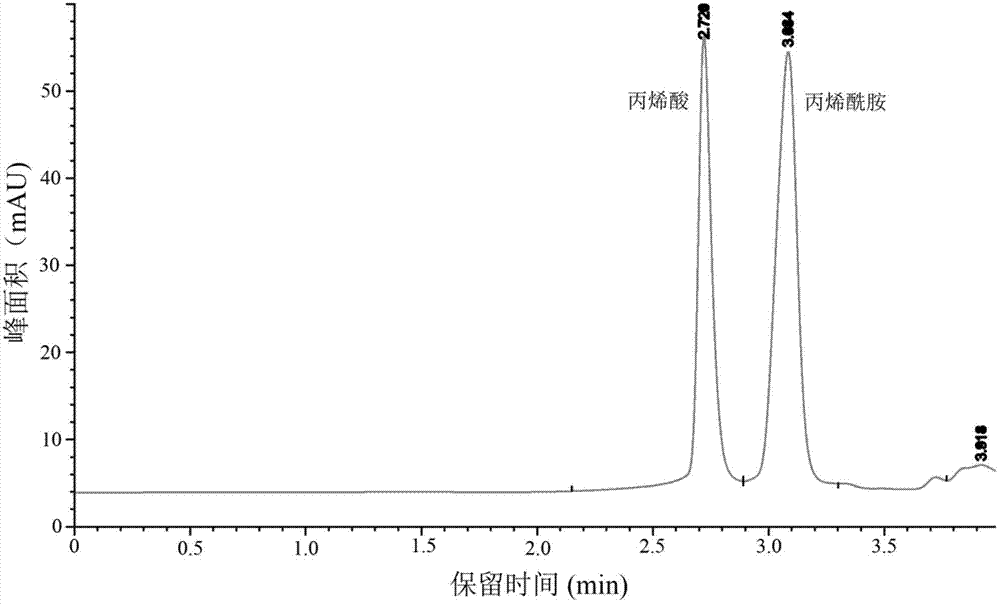
Amidase gene of Variovorax boronicumulans CGMCC 4969, and its application in biological degradation of acrylamide
The invention discloses Variovorax boronicumulans CGMCC 4969 capable of biologically degrading acrylamide. The amidase gene of the Variovorax boronicumulans CGMCC 4969 is composed of a DNA sequence represented by SEQ ID No:1 in a sequence table; DNA fragments composed of the sequence represented by the SEQ ID No:1 are inserted into an Escherichia coli expression vector pET28a plasmid to obtain a recombinant pET28a-amidase; and the recombinant pET28a-amidase is transformed into an Escherichia coli Rosetta strain, the amidase is expressed through induction, and Escherichia coli cells containing expression proteins can degrade acrylamide.
Owner:NANJING NORMAL UNIVERSITY
Composition and kit for detecting 23 respiratory pathogens and detection method of kit
PendingCN107488748AWith a high degree of automationStrong specificityMicrobiological testing/measurementMicroorganism based processesPhycoerythrinStreptavidin
The invention relates to a composition for detecting 23 respiratory pathogens. The composition comprises a primer composition consisting of the sequences shown by SEQ ID NO. 1-SEQ ID NO. 48 and a probe composition consisting of the sequences shown by SEQ ID NO. 49-SEQ ID NO. 72. The invention also relates to a kit for detecting 23 respiratory pathogens and a detection method of the kit. The kit comprises a first reaction solution, a second reaction solution, a first hybridization solution and a second hybridization solution containing the primers and probes respectively; the kit also comprises a mixed enzyme, a streptavidin-phycoerythrin solution, a negative reference, a positive reference and an interior label. The kit and the detection method provided by the invention integrate the functions of RT-PCR, full-automatic liquid-phase chip hybridization detection, data analysis and the like, and also have the advantages of high throughput, multiple detection target spots, high degree of automation, easiness in operation, short time consumption among the inventions of the same kind of liquid-phase chip technologies, etc.; moreover, the kit and the detection method provide important information for addressing the respiratory tract infection epidemic in time, and are of great significance to the prevention and control of respiratory pathogens.
Owner:SUZHOU SYM BIO LIFESCI CO LTD
Rice lesion mimic gene and encoded protein thereof
InactiveCN106243208AGood resistance to rice bacterial blightEasy to identifyPlant peptidesFermentationBiotechnologyNucleotide
The invention discloses a protein for controlling rice lesion mimic characters. The protein is formed by an amino acid sequence as shown by SEQ ID NO. 2 in a sequence table or a derivative sequence thereof. The invention further discloses a gene for encoding the protein. The gene is formed by a nucleotide sequence as show by SEQ ID NO. 1 in the sequence table. The lesion mimic gene provided by the invention has a good effect on resistance to rice bacterial blight, can be used for improving the bacterial blight resistance of nonglutinous rice, and provides a new way for breeding the nonglutinous rice capable of resisting bacterial blight; secondly, a new gene knockout technology is applied, a lesion mimic material under the background of japonica rice is acquired successfully, and a new method for utilizing the gene to culture japonica rice variety capable of resisting bacterial blight is provided. In addition, the gene provided by the invention is controlled by a single recessive gene, a transgene or filial generation can be simply identified and selected, and the convenience in application is realized.
Owner:SICHUAN AGRI UNIV
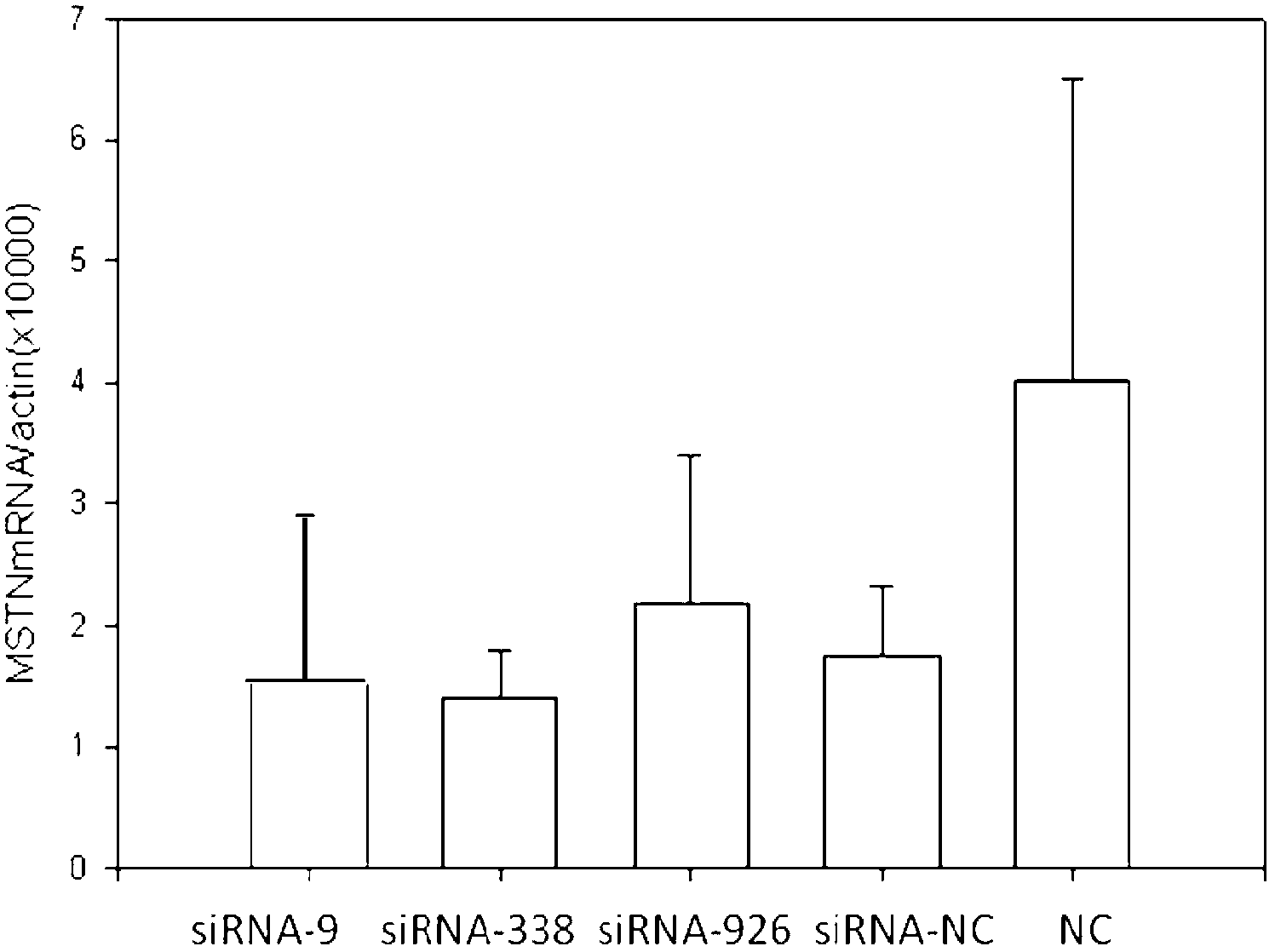
siRNA capable of restraining chicken myostatin gene expression and application thereof
ActiveCN102703449ASkeletal muscle enlargementIncrease meat productionMicroencapsulation basedAnimal husbandrySmall interfering RNABroiler
The invention discloses a siRNA capable of restraining chicken myostatin (MSTN) gene expression and application of the siRNA; small interfering RNA (ribonucleic acid) acted on myostatin gene is provided by the invention and is a) or b) or c) as follows: a) double-stranded RNA composed of a sequence 1 of a sequence table and a sequence 2 of the sequence table; b) double-stranded RNA composed of a sequence 3 and a sequence 4 of the sequence table; and c) double-stranded RNA composed of a sequence 5 and a sequence 6 of the sequence table; according to the invention, the small interfering RNA effectively capable of restraining chicken MSTN gene expression is obtained and is proved to effectively reduce expression of MSTN through real-time fluorescent quantitative PCR (polymerase chain reaction) on molecular biology; chicks with obviously enlarged skeletal muscle is successfully acquired through a micro-injection test on embryology; above-mentioned results show that the small interfering RNA provided by the invention can be used for breeding the chicks, so that chicken yield of the chicks specifically high-quality chicks is increased.
Owner:CHINA AGRI UNIV
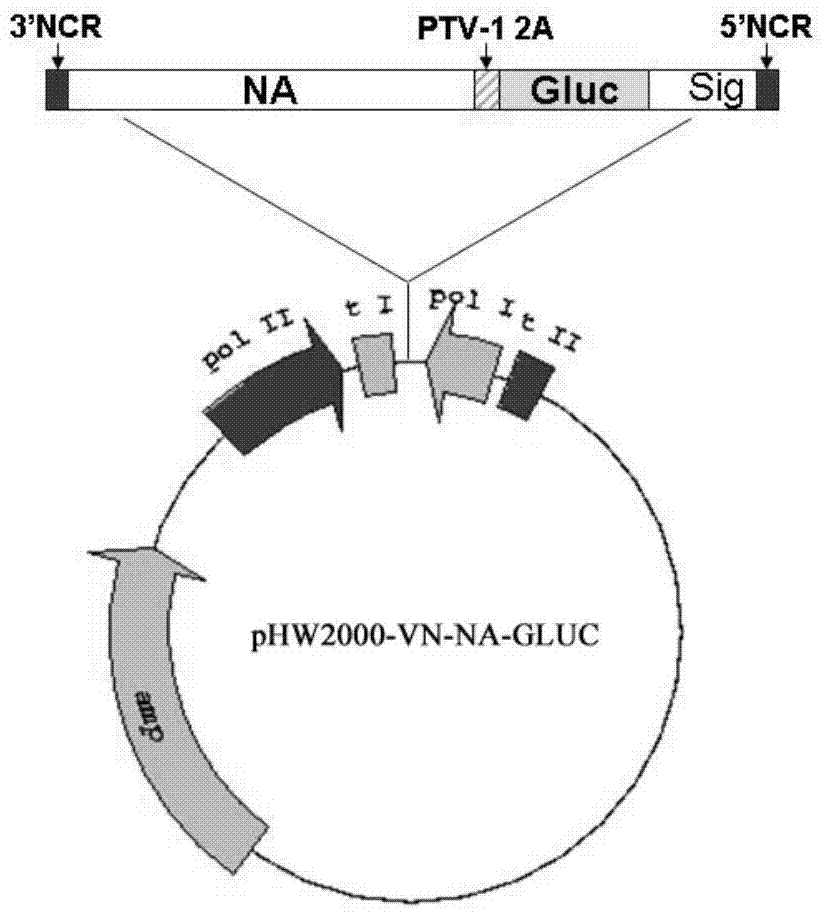
DNA fragment and application thereof in preparation of H5N1-subtype flu Guassia luciferase reporter virus
InactiveCN104513820ASimplify the titration procedureTrue Response Proliferation StatusViruses/bacteriophagesVector-based foreign material introductionSerum igeNucleotide
The invention discloses a DNA fragment and an application thereof in preparation of an H5N1-subtype flu Guassia luciferase reporter virus. The nucleotide sequence of the DNA fragment is composed of following sequences in a successive series manner: an H5N1-subtype flu virus NA segment 3'-terminal noncoded region coding gene sequence, an H5N1-subtype flu virus NA gene encoded region coding sequence, a PTV-1 virus 2A peptide encoded gene sequence, a Gaussia luciferase encoded gene sequence, a H5N1-subtype flu virus NA segment 5'-terminal packaged signal encoded gene sequence, and a H5N1-subtype flu virus NA segment 5'-terminal noncoded region coding sequence. By means of the recombined H5N1-subtype flu reporter virus prepared with the DNA fragments, quantitative detection on viruses in a sample can be objectively carried out quickly and accurately through activity of luciferase in cellular supernatant or animal serum. The H5N1-subtype flu Guassia luciferase reporter virus can not only save time but also improve accuracy and reliability of a detection result, thereby providing a new convenient and effective tool for drug screening and vaccine assessment of anti-H5N1-subtype flu virus drugs.
Owner:MICROBE EPIDEMIC DISEASE INST OF PLA MILITARY MEDICAL ACAD OF SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com