CRISPR detection primer group for mycoplasma pneumoniae and application of CRISPR detection primer group
A technology for detection of mycoplasma pneumoniae and primers, applied in the direction of microbe-based methods, microbiological measurement/testing, microbiology, etc., can solve the problem of inability to improve sensitivity, and achieve the goal of getting rid of the dependence on complex temperature-variable amplification instruments, high sensitivity, and broad The effect of applying the foreground
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
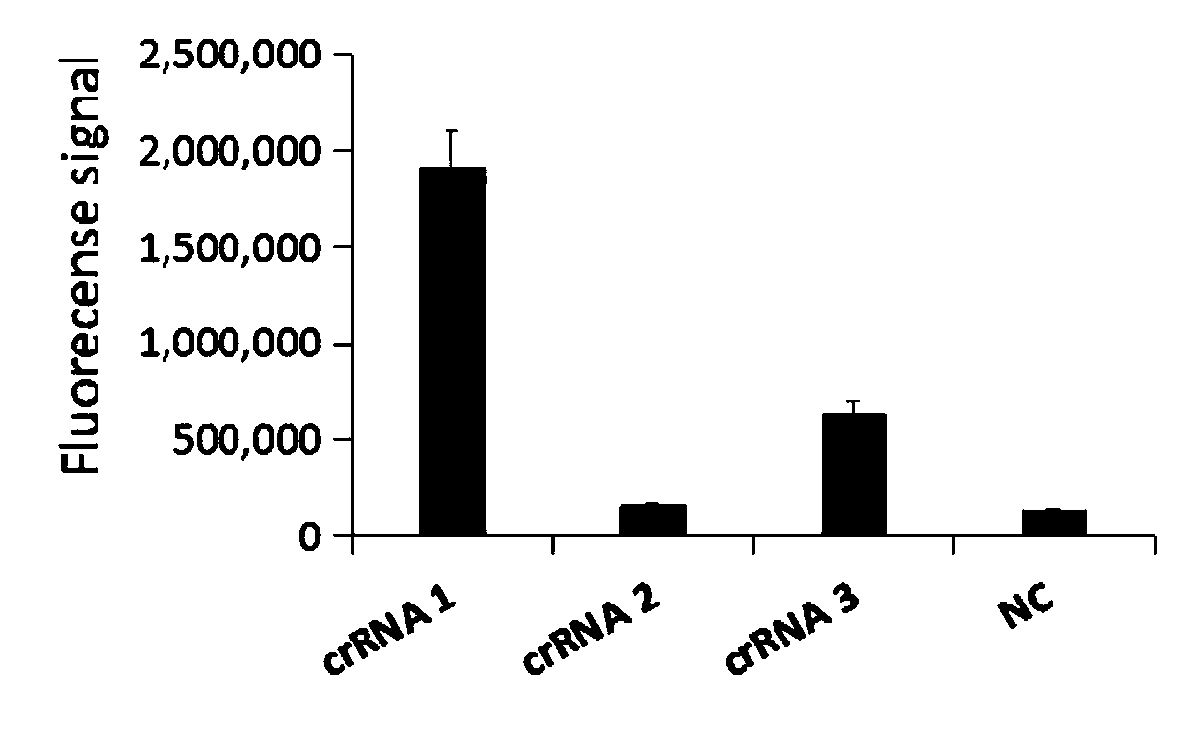
[0039] Design of primer sequences for CRISPR detection of Mycoplasma pneumoniae.
[0040] 1. Target sequence selection.
[0041] On the basis of the previous research, the inventors selected the P1 gene sequence region in the Mycoplasma pneumoniae genome that is specific and has no high-frequency SNP sites as the detection target sequence of Mycoplasma pneumoniae after multiple screenings and comparisons. The sequence of SEQ ID NO.1 is as follows:
[0042] 5’-TGATCTCGCCAACGCTCCCATTAAGCGGAGCGAGGAGTCGGGTCAGTCCGTCCAACTCAAGGCGGACGATTTTGGTACTGCCCTTTCCAGTTCGGGATCAGGCGGCAACTCCAATCCCGGTTCCCCCACCCCCTGAAGGCCGTGGCTTGCGACTGAGCAAATTCACAAGGACCTCCCCAAATGATCCGCCTCGATCCTGATTCTGTACGATGCGCCTTATGCGCGCAACCGTACCGCCATTGACCGCGTTGATCACTTGGATCCCAAGGCCATGACCGCGAACTATCCGCCCAGTTGAAGAACGCCCAAGTGAAACCACCACGGTTTGTGGGACTG-3’(SEQ ID NO.1)。
[0043] The standard sample to be tested in the following examples is the plasmid (synthesized by Aiji Biotechnology Co., Ltd.) inserted with the above-mentioned Mycoplasm...
Embodiment 2
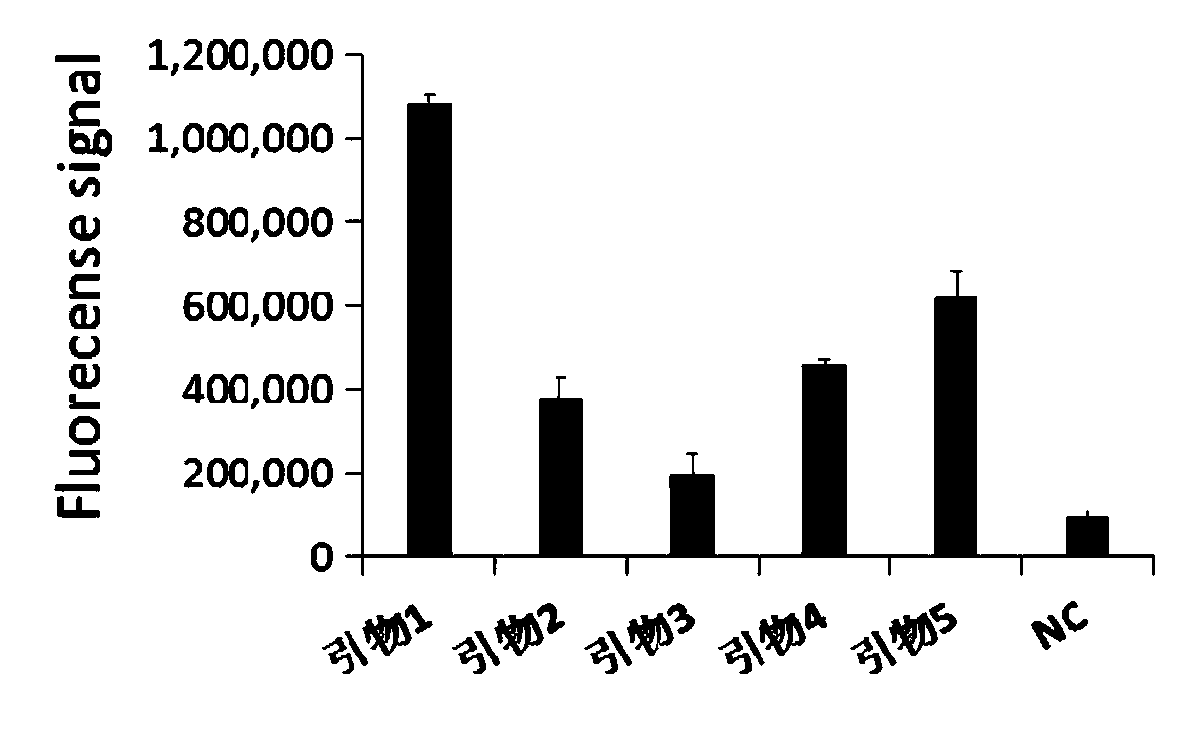
[0050] 1. The amplification efficiency screening of RPA amplification primers.
[0051] In order to screen the RPA amplification primer of Cas13a, insert the plasmid of the above-mentioned Mycoplasma pneumoniae P1 gene sequence in the above-mentioned embodiment 1 as the standard sample to be tested, extract the DNA group of the standard sample to be tested (i.e. the Mycoplasma pneumoniae genomic DNA) according to conventional methods, The screening primers include the above-mentioned primer pair 1, primer pair 2, primer pair 3, primer pair 4, and primer pair 5, wherein the template concentration is 1000 copies / μl.
[0052] 1.1 Method.
[0053] Amplify the target sequence by RPA, and use DEPC-treated water without impurity RNA, DNA and protein as a negative reference, select conventional reagents, and proceed as follows:
[0054] 1) RPA amplification
[0055] RPA reaction system is 50 μL: including RPA upstream primer 0.5-2.5 μL (concentration 10 μM), RPA downstream primer 0....
Embodiment 3
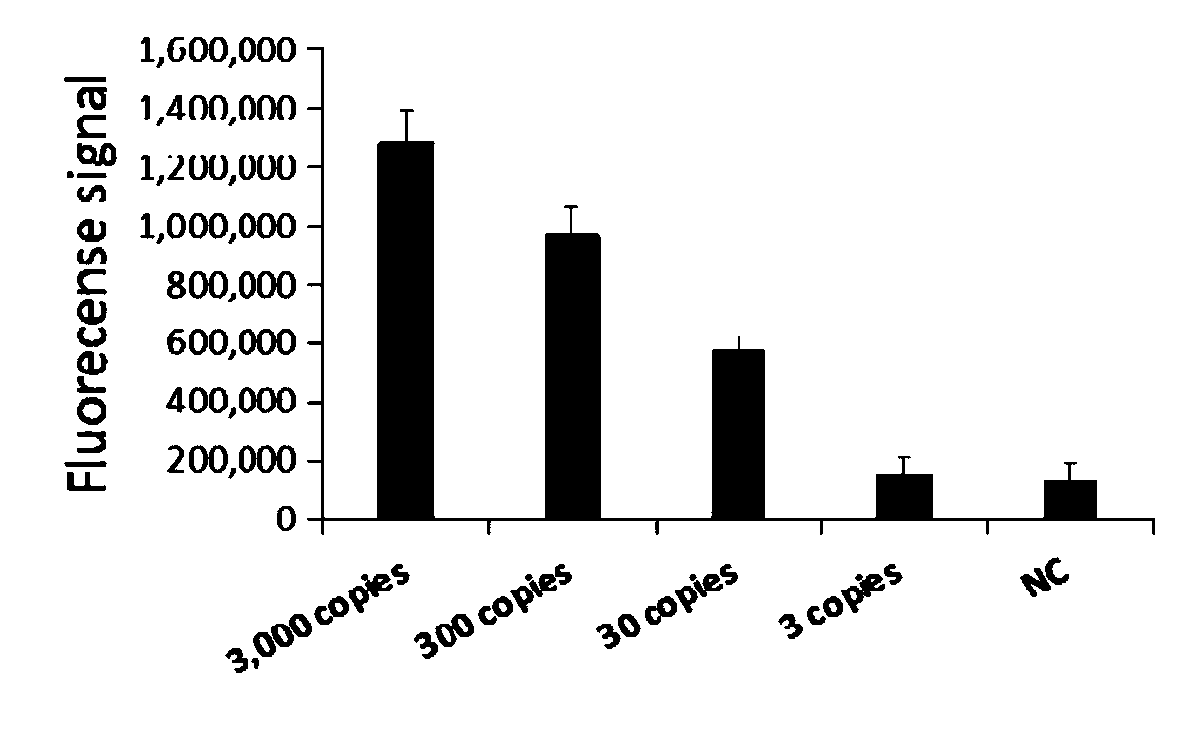
[0080] In this example, sensitivity detection is performed based on RPA amplification, T7 in vitro transcription and Cas13a.
[0081] The plasmid DNA inserted with the above-mentioned Mycoplasma pneumoniae P1 gene sequence was used as a template, and the calculated dilutions were 3000copies / μL, 300copies / μL, 30copies / μL, 3copies / μL, and 0copy / μL, a total of 5 gradients were used as templates for sensitivity detection.
[0082] 1. Method.
[0083] Referring to the method in Example 2 above, after RPA amplification, T7 transcription and CRISPR reaction were performed.
[0084] 2. Results.
[0085] ABI7500 fluorescence detector was used to detect the fluorescence of the amplified products. Among them, the result as image 3 As shown, it means that the detection limit of crRNA-1 is 30copies.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com