Circular RNA for translation in eukaryotic cells
A circular and nucleotide technology, applied in DNA/RNA fragments, recombinant DNA technology, nucleic acid vectors, etc., can solve problems such as short half-life
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
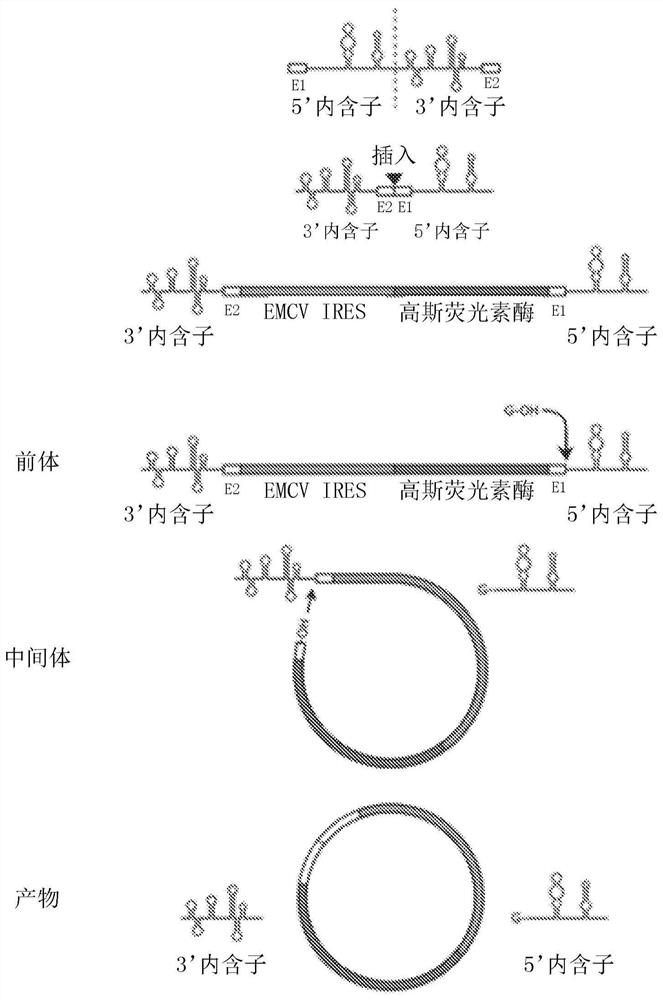
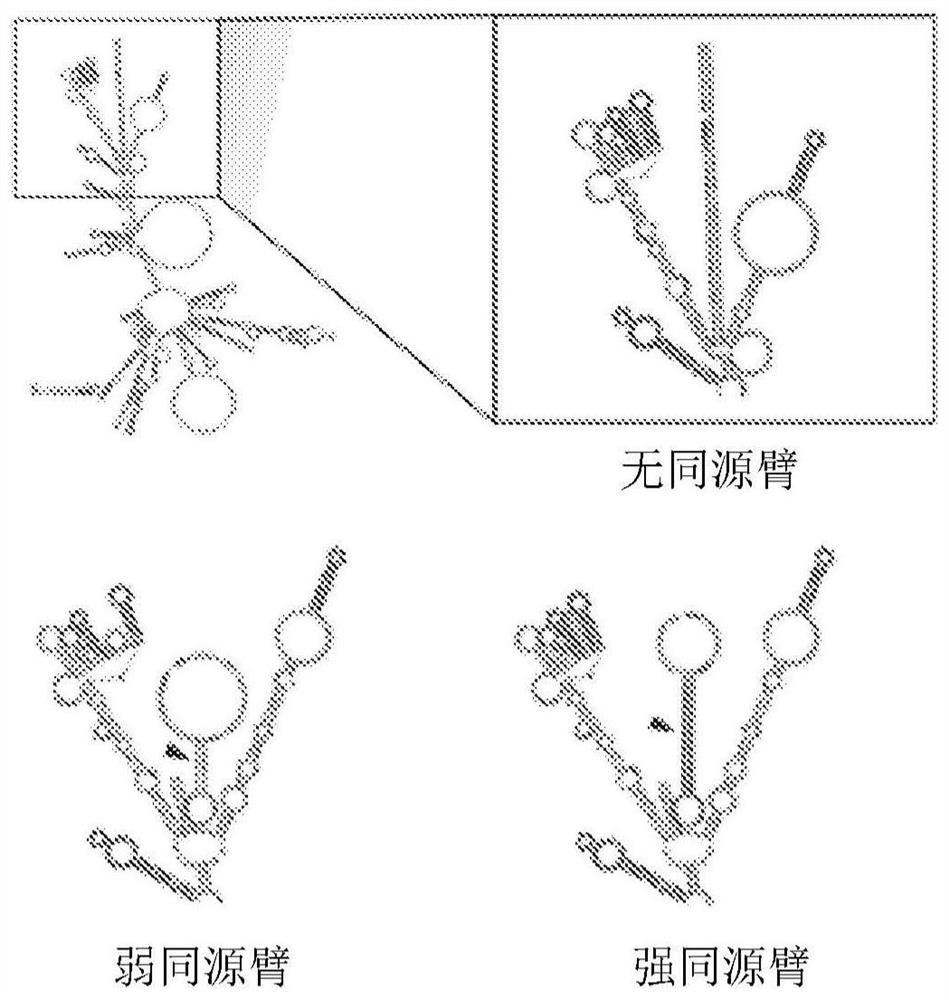
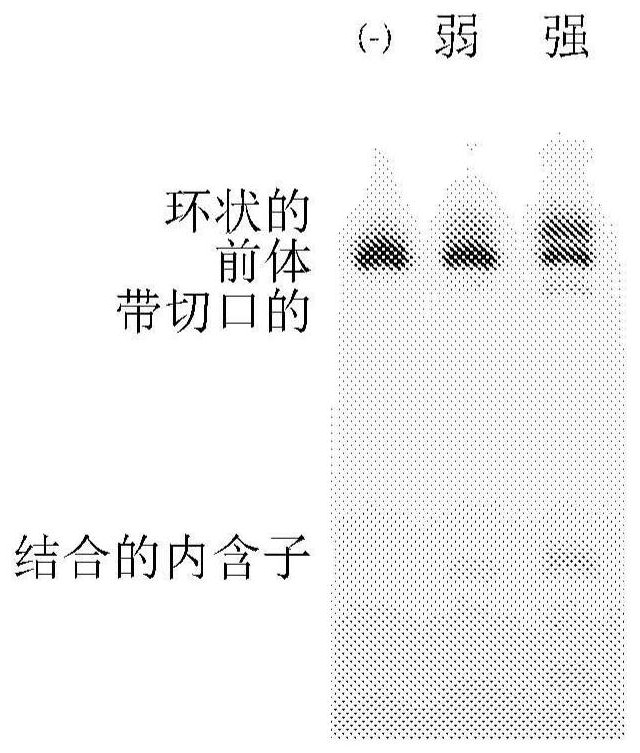
[0179] There are three general strategies for exogenous RNA circularization: chemical methods using cyanogen bromide or similar condensing agents, enzymatic methods using RNA or DNA ligases, and ribozyme methods using self-splicing introns (Petkovic, S. &Muller, S., "RNA circularization strategies in vivo and in vitro," Nucleic Acids Research, 43(4):2454-2465 (2015); Beadudry, D. & Perreault, J., "An efficient strategy for the synthesis of circular RNA molecules," Nucleic Acids Research, 23(15):3064-3066 (1995); Micura, R., "Cyclic Oligoribonucleotides (RNA) by Solid-Phase Synthesis," Chemistry-A European Journal, 5(7):2077- 2082 (1999)). It has been reported that the ribozyme method using the substituted group I catalytic intron is more suitable for long RNA circularization, requiring only the addition of GTP and Mg2+ as cofactors (Petkovic, S. & Muller, S., "RNA circularization strategies in vivo and in vitro," Nucleic Acids Research, 43(4):2454-2465(2015)). This permuted ...
Embodiment 2
[0215] Circular RNAs (circRNAs) are a class of single-stranded RNAs with continuous structure, enhanced stability and lack of terminal motifs necessary for interaction with various cellular proteins. Here we show that unmodified exogenous circRNAs are able to bypass cellular RNA sensors and thus avoid immune responses in RIG-1 and toll-like receptor (TLR) competent cells and mice. It was found that the immunogenicity and protein expression stability of circRNA preparations depended on the purity, and a small amount of contaminating linear RNA would lead to a strong cellular immune response. Unmodified circRNAs were less immunogenic than unmodified linear mRNAs in vitro, partly due to evading TLR induction and eliciting a cytokine response similar to that induced by uridine-modified linear mRNAs. Furthermore, uridine modification of circRNAs was found to disrupt internal ribosome entry site (IRES)-mediated translation and had no significant effect on cytokine responses. Finall...
PUM
| Property | Measurement | Unit |
|---|---|---|
| absorbance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com